Rationale and Objective
We sought to develop a Bayesian-filter that could distinguish positive radiology computed tomography (CT) reports of appendicitis from negative reports with no appendicitis.
Materials and Methods
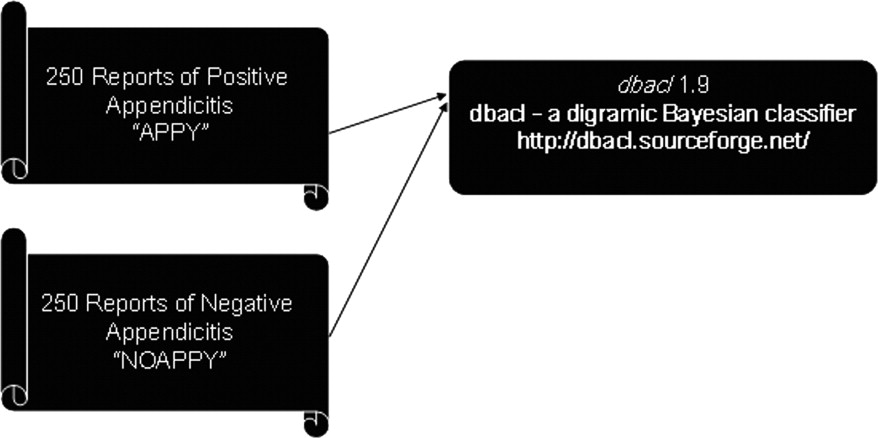
Standard unstructured electronic text radiology reports containing the key word appendicitis were obtained using a Java-based text search engine from a hospital General Electric PACS system. A total of 500 randomly selected reports from multiple radiologists were then manually categorized and merged into two separate text files: 250 positive reports and 250 negative findings of appendicitis. The two text files were then processed by the freely available UNIX-based software dbacl 1.9, a digramic Bayesian classifier for text recognition, on a Linux based Pentium 4 system. The software was then trained on the two separate merged text files categories of positive and negative appendicitis. The ability of the Bayesian filter to discriminate between reports of negative and positive appendicitis images was then tested on 100 randomly selected reports of appendicitis: 50 positive cases and 50 negative cases.
Results
The training time for the Bayesian filter was approximately 2 seconds. The Bayesian filter subsequently was able to categorize 50 of 50 positive reports of appendicitis and 50 of 50 reports of negative appendicitis, in less than 10 seconds.
Conclusion
A Bayesian-filter system can be used to quickly categorize radiology report findings and automatically determine after training, with a high degree of accuracy, whether the reports have text findings of a specific diagnosis. The Bayesian filter can potentially be applied to any type of radiologic report finding and any relevant category.
In the day-to-day practice of radiology, the ability to search prior reports for useful information is invaluable. The categorization and classification of radiology reports has been studied extensively ( ). A radiology picture archiving and communications system (PACS) environment contains a wealth of information that is not only useful for research purposes but also critical for quality assurance and improvement. However, as in most practices, radiology reports are unstructured and unique to the individual dictating the report. Rare cases, such as adrenal pheochromocytomas, can easily be found using the appropriate key words and Boolean operators and then manually sorting through the results ( ). These functions are provided by any number of commercial software packages. There are, however, limitations with this approach to data mining, which can be excessively time consuming.
For example, in a modern university-based radiology practice, a key word search for appendicitis returns over 10,000 results from a database of 1.2 million radiology reports. The reports may state, “There is no evidence of appendicitis” or “negative for appendicitis” or “consistent with appendicitis.” If a radiologist wanted to research the outcomes of patients with CT scans positive for appendicitis, he or she would be left to sort through a cumbersome and long list of results. Furthermore, refined Boolean search techniques do not aid the user in narrowing down the search results as the number of possibilities can be excessive ( ). There are simply too many ways in which CT-positive appendicitis can be expressed in a written report.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and methods
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Results
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusion
Get Radiology Tree app to read full this article<
References
1. Bijoy T., Hugue Q., Elkan H., Rosenthal D.: Automated computer-assisted categorization of radiology reports. AJR Am J Roentgenol 2005; 184: pp. 687-690.
2. Langer S.: OpenRIMS: An open architecture radiology informatics management system. J Digit Imaging 2002; 15: pp. 91-97.
3. Friedman C.: Towards a comprehensive medical language processing system: Methods and issues. Proc AMIA Annu Fall Symp 1997; pp. 595-599.
4. Friedman C.: A broad-coverage natural language processing system. Proc AMIA Symp 2000; pp. 270-274.
5. Spyns P.: Natural language processing in medicine: An overview. Methods Inf Med 1996; 35: pp. 285-301.
6. Huang Y., Lowe H.J.: A grammar-based classification of negations in clinical radiology reports. AMIA Annu Symp Proc 2005; pp. 988.
7. Haug P.J., Ranum D.L., Frederick P.R.: Computerized extraction of coded findings from free-text radiologic reports: Work in progress. Radiology 1990; 174: pp. 543-548.
8. Hripcsak G., Austin J.H., Alderson P.O., Friedman C.: Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology 2002; 224: pp. 157-163.
9. Chapman W.W., Haug P.J.: Comparing expert systems for identifying chest X-ray reports that support pneumonia. Proc AMIA Symp 1999; pp. 216-220.
10. Fiszman M., Chapman W.W., Evans S.R., Haug P.J.: Automatic identification of pneumonia related concepts on chest x-ray reports. Proc AMIA Symp 1999; pp. 67-71.
11. Chapman W.W., Cooper G.F., Hanbury P., Chapman B.E., Harrison L.H., Wagner M.M.: Creating a text classifier to detect radiology reports describing mediastinal findings associated with inhalational anthrax and other disorders. J Am Med Inform Assoc 2003; 10: pp. 494-503.
12. Jain N.L., Knirsch C.A., Friedman C., Hripcsak G.: Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proc AMIA Annu Fall Symp 1996; pp. 542-546.
13. Jain N.L., Friedman C.: Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp 1997; pp. 829-833.
14. Zdziarski J.A.: Tokenization: The building blocks of spam.Zdziarski J.A.Ending Spam: Bayesian Content Filtering and the Art of Statistical Language.2005.No Starch PressSan Francisco:pp. 96-109.
15. Bayesian spam filtering. Available online at: http://en.wikipedia.org/wiki/Bayesian_filtering . Accessed August 29, 2006.
16. Sahami M., Dumais S., Heckerman D., Horvitz E.: 1998.
17. A plan for spam. Available online at: http://www.paulgraham.com/spam.html . Accessed August 29,2006.
18. dbacl: A digramic Bayesian classifier. Available online at: http://dbacl.sourceforge.net . Accessed August 29, 2006.
19. Introduction to Bayesian filtering. Available online at: http://www.process.com/precisemail/bayesian_filtering.htm Accessed January 10, 2007.