Rationale and Objectives
Cancer screening by magnetic resonance imaging (MRI) has been shown to be one of the most sensitive methods available for the early detection of breast cancer. There is high variability in the diagnostic accuracy of radiologists analyzing the large amounts of data acquired in a breast MRI examination, and this has motivated substantial research toward the development of computer-aided detection and diagnosis systems. Most computer-aided diagnosis systems for breast MRI focus on dynamic information (how a lesion’s brightness changes over the course of an examination after the injection of a contrast agent). The inclusion of lesion margin measurements is much less common. One characteristic of malignant tumors is that they grow into neighboring tissues. This growth creates tumor margins that are variably fuzzy or diffuse (ie, they are not sharp).
Materials and Methods
In this short report, the authors present a new method for measuring a tumor’s margin from breast MRI examinations and compare it with an existing mathematical technique for margin measurements.
Results
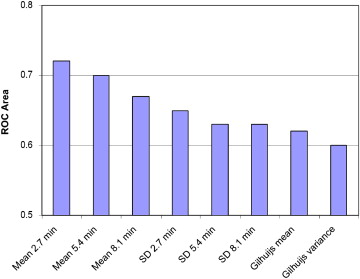
The proposed method can yield a test with sensitivity of 77% (specificity, 65%) on screening data, outperforming existing mathematical lesion margin measurement methods. Furthermore, when the presented margin measurement is combined with existing dynamic features, there is a statistically significant improvement in computer-aided diagnosis test performance ( P < .0014).
Conclusions
The proposed method for measuring a tumor’s margin outperforms existing mathematical methods on an extremely challenging data set containing many small lesions. The technique presented may be useful in discriminating between malignant and benign lesions in the context of the computer-aided diagnosis of breast cancer from MRI.
Genetic mutations on the BRCA1/2 genes can result in an 85% lifetime risk for developing breast cancer . Regular screening has been identified as key for improving survival rates . Dynamic contrast-enhanced magnetic resonance imaging (MRI) has been shown to be the most sensitive screening methodology for detecting breast cancer when compared with x-ray mammography, ultrasound, and clinical breast examination . It has also been demonstrated that in 3% of women diagnosed with unilateral breast cancer by mammography, pretreatment MRI examinations detect mammographically occult cancer in the contralateral breast . These studies suggest that MRI-based breast screening is likely to play a significant clinical role in the future. Furthermore, the American Cancer Society has recommended that women with a lifetime risk for developing breast cancer of 20% to 25% or greater should undergo MRI-based screening . It has been shown that there is a high degree of variability among trained radiologists in their ability to correctly diagnose lesions from breast MRI examinations . Breast MRI examinations typically involve the acquisition of hundreds of images, compared to just four images for typical x-ray mammographic screening. The data set used in this study is also abnormally challenging, containing lesions as small as 2 to 3 mm. This provides motivation for the research, design, and development of computer-aided detection and diagnosis systems to assist radiologists in identifying malignancies from these images.
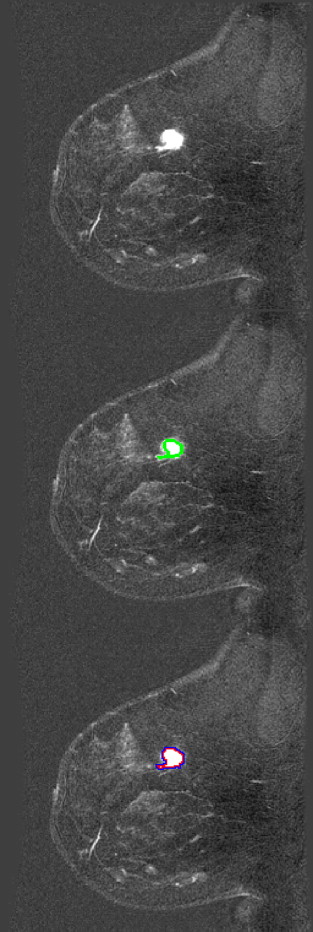
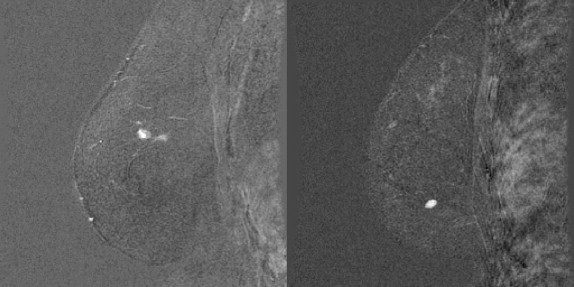
When analyzing a breast magnetic resonance image set, a radiologist will visually inspect the examination for a number of signs of malignancy. During the examination, the patient is imaged and then injected with a contrast agent, followed by more imaging. It is known that certain patterns in the changes in lesion brightness over time can be indicative of cancer, and this forms one of the main features that radiologists look for when making diagnoses from breast MRI examinations. Radiologists also look for spiculated lesions (or generally irregularly shaped lesions), heterogeneous tissue vascularization, and diffuse tumor edges, all of which are indicative of cancer and can affect their final diagnoses on the basis of the Breast Imaging Reporting and Data System reporting lexicon. Assessing tumor characteristics on the basis of the visual assessment of a radiologist is susceptible to human error, highlighting the need for automated methods for characterizing potentially malignant lesions. Although many methods have been presented in the literature for extracting various measurements of potential malignancy, relatively few methods have been developed for measuring the sharpness or diffuseness of a tumor’s margins, resulting in a gap in the literature on computer-aided diagnosis (CAD) for breast MRI. In this paper, we present a new method for measuring the sharpness and variability of a tumor’s margin.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and methods
Image Acquisition and Preprocessing
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Proposed Edge Sharpness Measurement
Get Radiology Tree app to read full this article<
RSIin(ri,t)−RSIout(n(ri),t)¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯d, RSI
in
(
r
i
,
t
)
−
RSI
out
(
n
(
r
i
)
,
t
)
¯
d
,
where RSI group is the relative signal intensity (normalized by the precontrast signal intensity) of a voxel in the inner or outer group, a function of position ( r i ) and time ( t ); r i is a voxel position marker in three-dimensional coordinates (the inner ring); n ( r i ) is the six-connect three-dimensional neighborhood operator, which provides a set of voxel positions that neighbor r i but are outside the lesion region of interest; t is the time point on which the margin measurement is being made (ie, a user parameter); t f is the final time point of the examination; and d is the normalization term RSIin(ri,t)−RSIout(n(ri),tf)¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯¯ RSI
in
(
r
i
,
t
)
−
RSI
out
(
n
(
r
i
)
,
t
f
)
¯ .
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Evaluation Technique: Existing Edge Sharpness Measurements
Get Radiology Tree app to read full this article<
Validation and Statistical Analyses
Get Radiology Tree app to read full this article<
Results
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
Acknowledgments
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Ford D., Easton D.F., Stratton M., et. al.: Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet 1998; 62: pp. 676-689.
2. Curry S.J.: Fulfilling the Potential of Cancer Prevention and Early Detection.2003.National Academies PressWashington, DC
3. Warner E., Plewes D.P., Hill K.A., et. al.: Surveillance of BRCA1 and BRCA2 mutation carriers with magnetic resonance imaging, ultrasound, mammography, and clinical breast examination. JAMA 2004; 292: pp. 1317-1325.
4. Lehman C., Gatsonis C., Kuhl C.K., et. al.: MRI evaluation of the contralateral breast in women with recently diagnosed breast cancer. N Engl J Med 2007; 356: pp. 1295-1303.
5. Pediconi F., Catalano C., Roselli A., et. al.: Contrast-enhanced MR mammography for evaluation of the contralateral breast in patients with diagnosed unilateral breast cancer or high-risk lesions. Radiology 2007; 243: pp. 670-680.
6. Lee S.G., Orel S.G., Woo I.J., et. al.: MR imaging screening of the contralateral breast in patients with newly diagnosed breast cancer: preliminary results. Radiology 2003; 226: pp. 773-778.
7. Saslow D., Boetes C., Burke W., et. al., American Cancer Society Breast Cancer Advisory Group: American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. Cancer J Clin 2007; 57: pp. 75-89.
8. Warren R., Hayes C., Pointon L., et. al.: A test of performance of breast MRI interpretation in a multicentre screening study. Magn Reson Imaging 2006; 24: pp. 917-929.
9. Gilhuijs K., Giger M.L., Bick U.: Computerized analysis of breast lesions in three dimensions using dynamic magnetic-resonance imaging. Med Phys 1998; 25: pp. 1647-1654.
10. Gilhuijs K., Giger M.L., Bick U.: Automated feature extraction and classification of breast lesions in magnetic resonance images. Proc SPIE 1998; 3338: pp. 294-300.
11. Penn A., Thompson S., Brem R., et. al.: Morphologic blooming in breast MRI as a characterization of margin for discriminating benign from malignant lesions. Acad Radiol 2006; 13: pp. 1344-1354.
12. Greenman R.L., Lenkinski R.E., Schnall M.D., et. al.: Bilateral imaging using separate interleaved 3D volumes and dynamically switched multiple receive coil arrays. Magn Reson Med 1998; 39: pp. 108-115.
13. Martel A.L., Froh M.S., Brock K.K., et. al.: Evaluating an optical-flow-based registration algorithm for contrast-enhanced magnetic resonance imaging of the breast. Phys Med Biol 2007; 52: pp. 3803-3816.
14. Levman J., Causer P., Warner E., et. al.: Effect of the enhancement threshold on the computer-aided detection of breast cancer using MRI. Acad Radiol 2009; 16: pp. 1064-1069.
15. Eng J.: Receiver operating characteristic analysis: a primer. Acad Radiol 2005; 12: pp. 909-916.
16. Levman J., Leung T., Causer P., et. al.: Classification of dynamic contrast-enhanced magnetic resonance breast lesions by support vector machines. IEEE Trans Med Imaging 2008; 27: pp. 688-696.