Rationale and Objectives
RadLex is a standardized vocabulary developed for clinical practice, research, and education in radiology. This report sought to analyze the use of RadLex to annotate and index the captions of images from the peer-reviewed biomedical literature and to compare the number of annotations per term for RadLex and five other biomedical ontologies in a large corpus of figure captions from biomedical imaging publications.
Materials and Methods
RadLex and five other biomedical vocabularies were evaluated. A fully automated web service was used to discover the vocabularies’ terms in a collection of 385,018 figure captions from 613 peer-reviewed biomedical journals. Annotations (i.e., figure-term pairs) were analyzed by vocabulary. RadLex annotations were analyzed by journal and RadLex term class.
Results
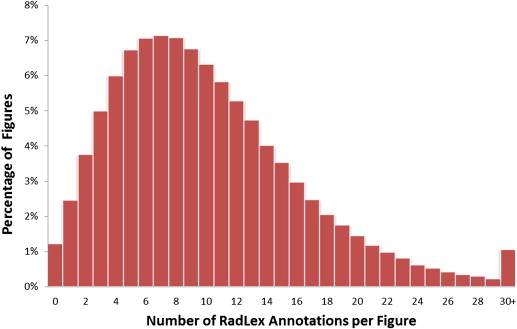
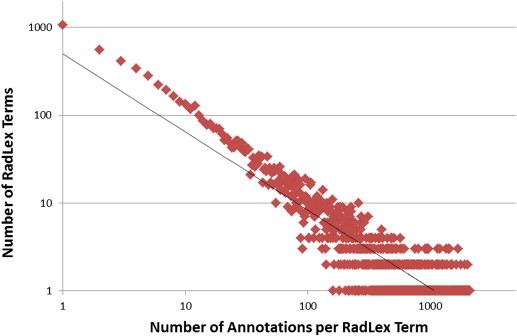
RadLex had the greatest number of annotations per term of the six vocabularies. On average, there were 10.1 RadLex annotations per figure; 380,338 figures (98.8%) were annotated with at least one RadLex term and 288,163 figures (74.8%) were annotated with six or more RadLex terms. Of 39,218 RadLex terms, 8504 (21.7%) were mapped to images in the collection, which was the highest percentage of any of the vocabularies.
Conclusions
Although comprising four to 10 times fewer terms than other vocabularies, RadLex showed excellent performance in indexing radiology-centric content. Almost all of the images in a large collection of figures from peer-reviewed biomedical journals were annotated with at least one RadLex term, and almost 75% of the images were annotated with six or more terms.
The RadLex radiology lexicon ( www.radlex.org ) is a collection of terms designed to provide a uniform vocabulary of radiology . It has been developed under the aegis of the Radiological Society of North America and plays an increasingly important role in radiology practice, research, and education. RadLex is being used to categorize journal articles and reviewers , encode radiology result information , search the content of radiology reports , analyze queries to web-based search engines , and standardize names of imaging procedures for the American College of Radiology’s Dose Index Registry .
RadLex provides an ontology (knowledge model) that incorporates relationships between terms . The main relationship is the subsumption (“is-a”) relation, which defines a hierarchy of subclasses. For example, the term “left lung” RadLex term identifier (RID1326) is a subclass of “lung” (RID1301), and “bronchitis” (RID34637) is a subclass of “respiratory disorder” (RID5316). In RadLex, each entity is related to a single, higher level entity, or “parent.” This hierarchical representation of radiology terms allows retrieval of information more effectively.
Get Radiology Tree app to read full this article<
Materials and methods
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 1
Comparison of Six Ontologies for Annotation of the ARRS GoldMiner Corpus of Figure Captions Using the Most Recent Version of Each Ontology at the NCBO BioPortal Site
Ontology Version Release Date No. of Terms Annotated Terms Annotated Figures Annotations Number Percent Number Percent Number Per Term Per Figure FMA 3.1 March 3, 2010 83,281 5398 6.5 324,376 84.2 1,288,568 15.5 3.3 ICD-10-CM 2011_01 January 1, 2011 91,590 1635 1.8 84,987 22.1 104,095 1.1 0.3 LOINC 236 June 1, 2011 171,399 7683 4.5 380,834 98.9 5,008,536 29.2 13.0 MeSH 2012 September 9, 2011 229,698 15,792 6.9 381,978 99.2 3,097,452 13.5 8.0 RadLex 3.8 February 19, 2013 39,218 8504 21.7 380,338 98.8 3,871,573 98.7 10.1 SNOMED CT 2011_07_31 July 31, 2011 395,036 41,371 10.5 384,492 99.9 11,588,578 29.3 30.1 Total 1,010,222 80,383 8.0 385,018 24,958,802 24.7 64.8
FMA, Foundational Model of Anatomy; ICD-10-CM, International Classification of Diseases, Version 10, Clinical Modification; LOINC, Logical Observation Identifier Names and Codes; MeSH, Medical Subject Headings; SNOMED CT, Systematized Nomenclature of Medicine–Clinical Terms.
The Annotated Terms column shows the number of terms from each ontology that appeared in the annotations. The Annotated Figures column shows the number of figures captions from the collection that were annotated.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Results
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
t=499.69⋅a−0.889. t
=
499.69
⋅
a
−
0.889
.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 2
Frequency of Annotations for Major RadLex Classes
Major RadLex Class Number of Terms Terms with Annotations Mean Number of Annotations per Term Number Percent Anatomical entity 32,728 4458 14 30 Clinical finding 2146 1587 74 247 Imaging modality 55 46 84 1717 Imaging observation 932 400 43 277 Imaging procedure attribute 878 322 37 81 Medical device 325 168 52 126 Nonanatomical substance 366 205 56 75 Object 45 28 62 43 Procedure 423 278 66 397 Procedure step 97 57 59 451 Process 6 6 100 1086 Property 234 153 65 801 RadLex descriptor 886 742 84 1598 RadLex nonanatomical set 4 0 0 — Report component 18 11 61 1157 Total 39,143 8461 22 98
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 3
RadLex Figure Annotations by Source
Source Number of Figures Number of Words Mean Words per Figure Number of RadLex Annotations Mean RadLex Annotations per Figure_American Journal of Neuroradiology_ 16,210 1,284,355 79.2 215,125 13.3American Journal of Roentgenology 77,998 3,954,639 50.7 902,100 11.6British Journal of Radiology 8831 500,368 56.7 90,346 10.2 Eurorad 22,220 648,241 29.2 191,269 8.6Journal of Nuclear Medicine 13,300 887,357 66.7 99,432 7.5RadioGraphics 51,841 3,143,696 60.6 540,659 10.4Radiology 58,792 3,173,463 54.0 656,346 11.2 “Core” sources 249,192 13,592,119 54.5 2,695,277 10.8 Other sources 135,826 10,329,648 76.1 1,176,296 8.7 Total 385,018 23,921,767 62.1 3,871,573 10.1
The seven “core” radiology sources accounted for almost two-thirds of all figures. Although they had fewer words per figure, they had a greater number of RadLex annotations per figure.
Table 4
The 20 Most Frequently Annotated RadLex Terms
RadLex Identifier Term Name (Synonyms) Number of Figures Annotated RID5824 Left 71,190 RID39330 After 68,807 RID5825 Right 68,309 RID10312 Magnetic resonance imaging (MRI) 39,034 RID39055 Breast mass (mass; nodule) 35,150 RID478 Artery 33,224 RID3874 Mass 30,349 RID39466 Liver mass (mass) 30,349 RID39056 Lung mass (mass) 30,349 RID34373 Mass in or on skin (mass) 30,349 RID13173 Normal 28,438 RID5861 Coronal 28,190 RID3957 Neoplasm 26,234 RID8 Treatment 26,014 RID38780 Lesion 24,638 RID5818 Anterior 23,676 RID34616 Malignant neoplastic disease (cancer) 21,393 RID39348 Cell 21,034 RID4247 Carcinoma 21,034 RID5860 Sagittal 20,943
Note that all of these terms consist of a single word or have a single-word synonym, which likely accounts for their high frequency in the figure captions. Three terms that have the synonym “mass” (RID39466, RID39056, and RID34373) have the same number of annotations as term RID3874 (“mass”).
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Appendix
Get Radiology Tree app to read full this article<
Major Class Terms with the Most Annotations Terms without Annotations (randomly selected) No. of Figures Annotated Term Term anatomical entity 33,224 artery Brodmann area 13 of left temporal lobe 21,034 cell hyaloid canal 18,497 brain long ciliary nerve to left ciliary body 18,337 liver medial part of left medial mammillary nucleus 17,752 wall muscle body of right pronator teres 16,500 human region of prostatic sinus 16,233 portion of tissue subaortic common iliac lymph node 16,175 lung surface of left occipital lobe 15,534 thorax tendon of left extensor digitorum to left little finger 14,111 breast thoracic subsegment of dorsal gray column of spinal cord clinical finding 26,234 neoplasm aspiration of digestive secretions 21,393 malignant neoplastic disease complicated cysts 21,034 carcinoma congenital agammaglobulinemia 14,311 pain congenital anomaly of spine 10,743 stenosis disorder caused by drugs or toxins 9,468 obstruction focal cortical dysplasia without balloon cells 7,999 infarction fracture healing disorder 7,581 thickening injury due to thermal or electrical trauma 6,508 aneurysm melanocytic medulloblastoma 6,389 hemorrhage type 2 endplate marrow change imaging modality 39,034 magnetic resonance imaging conventional tomography 14,295 ultrasound high intensity pulse 11,215 tomography high-energy scan 5,713 computed tomography low intensity pulse 5,585 projection radiography low-energy scan 2,723 mammography panographic radiograph 2,477 spectroscopy phase-cancellation harmonic ultrasound 2,234 positron emission tomography planar nuclear medicine imaging 1,923 scintigraphy single photon imaging 1,518 functional magnetic resonance imaging imaging observation 35,150 breast mass cockade image 30,349 liver mass deflected calyx sign 30,349 lung mass hilum convergence sign 30,349 mass lace-like pattern of joint 30,349 mass in or on skin paradoxical halo sign 24,638 lesion pelvocalyceal wall opacification sign 19,871 enhancement popcorn calcification sign 13,683 assessment spatial enhancement pattern 10,816 cirrhosis-associated nodules waterfall hilum 5,504 enhancing yo-yo-on-a-string sign imaging procedure attribute 8,321 multi-detector arrhythmia artifact 5,518 radiographic projection closure device access closure technique 4,181 regression dual interpretation 3,978 arterial phase duration of ablation endpoint 3,535 fast-spin echo gaynor hart view 3,008 flip angle immediate complication 2,836 artifact microscopic motion artifact 2,655 fluid-attenuated inversion recovery response to non-vascular treatment 1,973 portal venous phase settegast view 1,919 t1 weighted wrong demographics medical device 5,203 continuously rotating tube ct ACL graft device 5,153 valve biopsy coil 4,957 stent cryoplasty balloon 4,623 catheter hybrid fixator 3,741 needle low-pressure balloon 2,835 tube monopolar applicator 2,125 balloon non-linear shim coil 1,202 electrode nuclear imaging device 1,106 prosthesis temporary pacemaker patch 1,091 filter xenon ionization chamber non-anatomical substance 3,032 gadolinium americium 2,571 contrast agent I-123 carcinoembryonic antigen 1,689 iron ionic low-osmolality dimeric iodinated contrast agent 1,633 calcium Meitnerium object 457 foreign body bevel needle tip 377 needle tip diamond needle tip 225 adhesive flexible diffuser laser tip 216 knife Hemostatic sheath 134 bullet medical object 134 intrauterine device personal item 46 coin platinum embolization coil 45 bezoar serrated needle tip 41 embolic material steerable wire tip 33 shrapnel straight wire tip procedure 26,014 treatment three phase imaging 17,989 angiography nuchal translucency evaluation 11,468 follow-up procedure obtain access 8,909 surgical procedure ultrasound-assisted venipuncture 8,630 biopsy platelet-rich plasma injection 5,969 ablation extraction of biliary calculi 5,751 diffusion imaging with iv contrast 5,669 echocardiography nephrotomography 5,342 placement imaging without iv contrast 4,724 screening small bowel transplantation procedure step 18,647 mean value calculation counts per pixel calculation 5,720 average value calculation diffusion tensor reconstruction 4,391 maximum intensity projection hardware image fusion with reregistration 3,155 apparent diffusion coefficient map image-based parallel imaging reconstruction 1,932 intensity projection iterative partial fourier reconstruction 1,669 multiplanar reformat k-space filtering 1,143 volume rendering primary reconstruction step 967 calculation reformat procedure step 853 cerebral blood flow map trace of diffusion tensor reconstruction 749 tractography zero-filling of 3d volume k-space data process 3,900 motion 1,238 metabolism 1,163 gestation 146 swallowing 67 peristalsis 4 biological process property 16,785 diagnosis 10 o’clock position 15,024 flow 2 o’clock position 13,832 volume 3 o’clock position 13,374 phase arm pedaling exercise 11,143 function diffusely increased vascularity in surrounding tissue 7,235 measurement intermediate perceptual difficulty 7,156 color mimics another entity 6,453 maximum rolled medial 6,277 location t2 tumor stage 6,136 typical vascular flow measurement mode RadLex descriptor 71,190 left aortic dissection modifier 68,807 after body region covered 68,309 right disc containment 28,438 normal displaces portal vein 28,190 coronal extra-amniotic 23,676 anterior hormonal pathophysiologic process modifier 20,943 sagittal intrameningeal 20,098 transverse polyp morphology characteristic 19,974 small quality descriptor 19,888 both turbidity modifier
Get Radiology Tree app to read full this article<
References
1. Langlotz C.P.: RadLex: a new method for indexing online educational materials. RadioGraphics 2006; 26: pp. 1595-1597.
2. Klein J.S.: A look back at 2012 and plans for 2013. RadioGraphics 2013; 33: pp. 1-2.
3. Kahn C.E., Langlotz C.P., Burnside E.S., et. al.: Toward best practices in radiology reporting. Radiology 2009; 252: pp. 852-856.
4. Channin D.S., Mongkolwat P., Kleper V., et. al.: The annotation and image mark-up project. Radiology 2009; 253: pp. 590-592.
5. Lacson R., Andriole K.P., Prevedello L.M., et. al.: Information from Searching Content with an Ontology-Utilizing Toolkit (iSCOUT). J Digit Imaging 2012; 25: pp. 512-519.
6. Rubin D.L., Flanders A., Kim W., et. al.: Ontology-assisted analysis of web queries to determine the knowledge radiologists seek. J Digit Imaging 2011; 24: pp. 160-164.
7. Morin R.L., Coombs L.P., Chatfield M.B.: ACR Dose Index Registry. J Am Coll Radiol 2011; 8: pp. 288-291.
8. Rubin D.L.: Creating and curating a terminology for radiology: ontology modeling and analysis. J Digit Imaging 2008; 21: pp. 355-362.
9. Kahn C.E., Thao C.: GoldMiner: a radiology image search engine. AJR Am J Roentgenol 2007; 188: pp. 1475-1478.
10. Rosse C., Mejino J.L.: A reference ontology for biomedical informatics: the foundational model of anatomy. J Biomed Inform 2003; 36: pp. 478-500.
11. Barta A., McNeill G., Meli P., et. al.: ICD-10-CM primer. J AHIMA 2008; 79: pp. 64-66. quiz 67–68
12. Huff S.M., Rocha R.A., McDonald C.J., et. al.: Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc 1998; 5: pp. 276-292.
13. McDonald C.J., Huff S.M., Suico J.G., et. al.: LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003; 49: pp. 624-633.
14. Medical Subject Headings. National Library of Medicine; 2007. Available at: http://www.nlm.nih.gov/mesh/ . Accessed October 3, 2007.
15. 2012.International Health Terminology Standards Development OrganisationCopenhagen
16. Musen M.A., Noy N.F., Shah N.H., et. al.: The National Center for Biomedical Ontology. J Am Med Inform Assoc 2012; 19: pp. 190-195.
17. Whetzel P.L., Noy N.F., Shah N.H., et. al.: BioPortal: enhanced functionality via new web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res 2011; 39: pp. W541-W545.
18. Bodenreider O.: The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32: pp. D267-D270.
19. Jonquet C., Shah N.H., Musen M.A.: The open biomedical annotator. Summit Translat Bioinform 2009; pp. 56-60.
20. Langlotz C.P., Caldwell S.A.: The completeness of existing lexicons for representing radiology report information. J Digit Imaging 2002; 15: pp. 201-205.
21. Aronson A.R.: Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001; pp. 17-21.
22. Kahn C.E., Rubin D.L.: Improving radiology image retrieval through automated semantic indexing of figure captions. J Am Med Informatics Assoc 2009; 16: pp. 370-376.
23. Dai M, Shah NH, Xuan W, et al. An efficient solution for mapping free text to ontology terms. AMIA Summit on Translational Bioinformatics. San Francisco, CA; 2008.
24. Shah N.H., Bhatia N., Jonquet C., et. al.: Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinform 2009; 10: pp. S14.
25. Rubin D.L., Shah N.H., Noy N.F.: Biomedical ontologies: a functional perspective. Brief Bioinform 2008; 9: pp. 75-90.
26. Bodenreider O.: Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearbk Med Inform 2008; pp. 67-79.
27. Musen M.A., Shah N.H., Noy N.F., et. al.: BioPortal: ontologies and data resources with the click of a mouse. AMIA Annu Symp Proc 2008; pp. 1223-1224.
28. Smith B., Ashburner M., Rosse C., et. al.: The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 2007; 25: pp. 1251-1255.
29. Tao C., Pathak J., Solbrig H.R., et. al.: Terminology representation guidelines for biomedical ontologies in the semantic web notations. J Biomed Inform 2013; 46: pp. 128-138.
30. Shah N.H., Jonquet C., Chiang A.P., et. al.: Ontology-driven indexing of public datasets for translational bioinformatics. BMC Bioinform 2009; 10: pp. S1.