Objectives
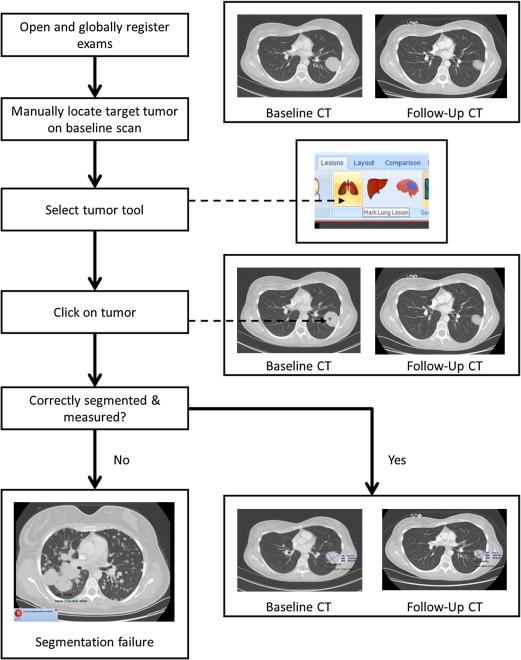
Our goal was to evaluate a new software capability that integrates registration, segmentation and tumor measurement across serial exams within a picture archiving communication system (PACS) to expedite tumor measurement.
Materials and Methods
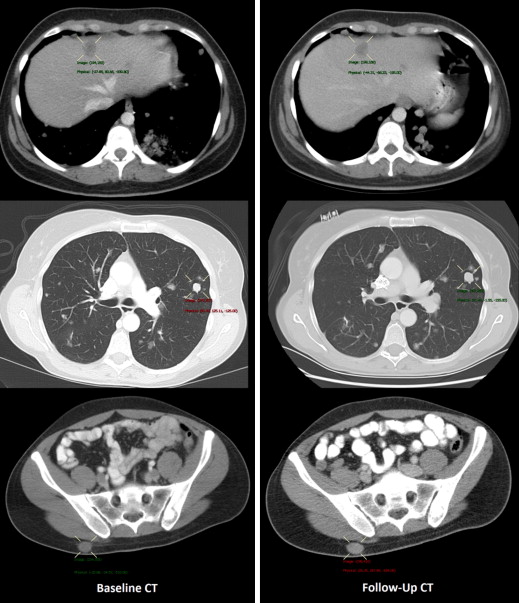
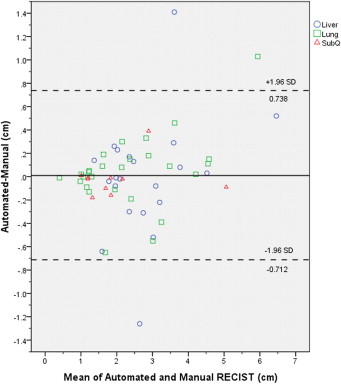
Patients treated under institutional review board–approved protocols for metastatic melanoma were retrospectively reviewed. Of the 19 included patients, five were male, the median age was 43.2, and all received treatment using an adoptive cell therapy. Seventy-one lung, liver, and subcutaneous tumors were manually measured using RECIST (Response Evaluation Criteria In Solid Tumors) criteria before therapy (baseline computed tomography [CT]) and within 3 months after therapy (follow-up CT). We performed semiautomated registration, segmentation, and RECIST measurements at both time points within PACS (Carestream Health, Rochester, NY). We compared manual and software-generated RECIST measurements using Bland-Altman plots.
Results
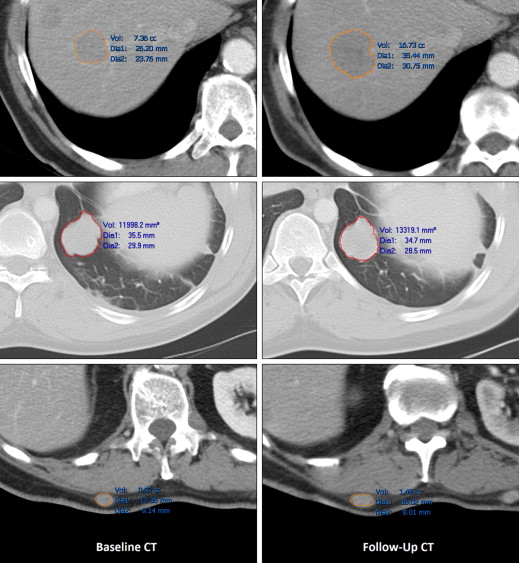
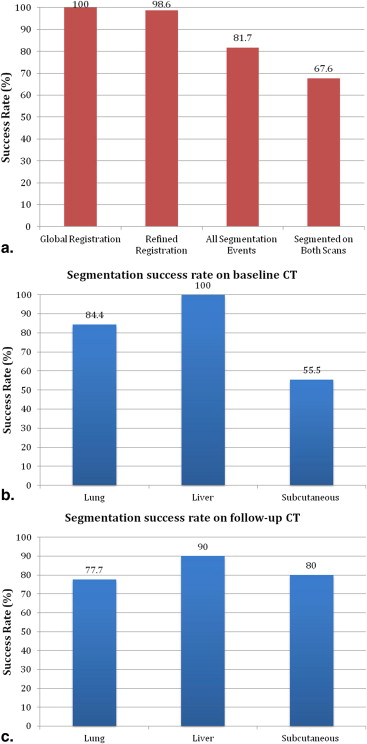
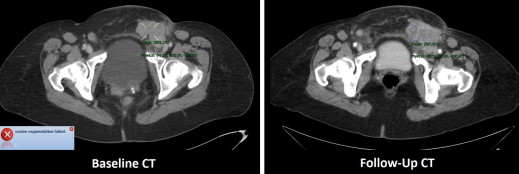
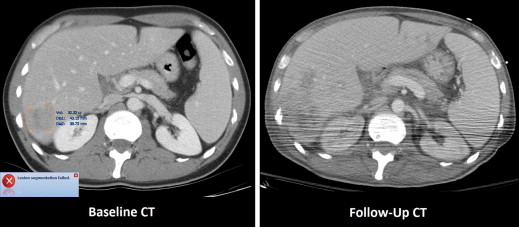
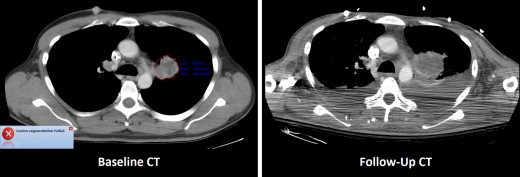
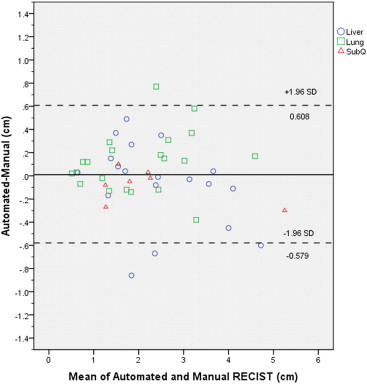
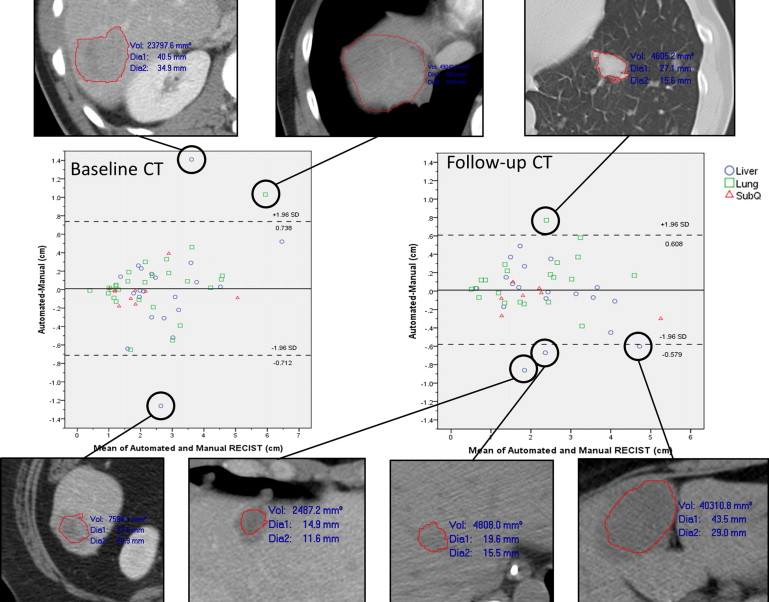
The median manually measured RECIST diameter for all baseline tumors was 2.1 (1.0–6.2) cm. The refined registration function identified 70/71 (98.6%) tumors on the follow-up CT. On the baseline CT, all 21 liver, 27/32 (84%) lung, and 10/18 (55%) subcutaneous tumors completed segmentation. On the follow-up CT, 19/21 (90%) liver, 21/27 (78%) lung, and 8/10 (80%) subcutaneous tumors completed segmentation. The Bland-Altman plot demonstrated a 95% confidence interval of ±0.7 cm when comparing the software-generated and manual RECIST measurements.
Conclusions
The PACS software performed semiautomated baseline tumor measurements and fully automated follow-up tumor measurements in a majority of lung, liver, and subcutaneous tumors. In our patients, semiautomated metastatic tumor measurement did not obviate the need for physician oversight due to disease and treatment-related factors.
Metastatic tumors are identified and measured on serial computed tomography (CT) scans to evaluate the efficacy of cancer treatment. The current method of metastatic tumor evaluation by CT scan is called the Response Evaluation Criteria In Solid Tumors (RECIST) . Manual measurement of metastatic tumors using RECIST criteria often consumes significant time for radiologists and treatment teams . In a survey of academic radiologists working at National Cancer Institute–funded cancer centers, 87% responded that they perform some tumor measurements in the first clinical scan, but only 7% provide measurement of all tumors included in the report . In the same study, 86% agreed they would include tumor measurements in reports if the measurements could be completed with one click of a mouse. Semiautomated software-generated RECIST measurements have recently developed in an effort to improve efficiency and allow more widespread inclusion of RECIST measurements in radiology reports . We describe the software applied in this study below that semiautomatically measures metastatic tumors on serial CT scans using minimal manual input and a one-click fully automated tumor measurement on serial CT exams.
Software-generated recist measurement
Software-generated tumor localization in serial CT scans relies on recent advances in image processing including registration and tumor segmentation. Software algorithms facilitate the registration of planes between two CT scans, for example, by minimizing the square differences between the datasets . Segmentation employs enhanced tumor edge detection and other methods to distinguish tumors from surrounding structures . Software-based systems often couple segmentation and tumor measurement, providing axial tumor length, cross-sectional area, and tumor volume .
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and methods
Patient Population
Get Radiology Tree app to read full this article<
CT Scans and Manual Measurements
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Semiautomated Registration, Segmentation, and RECIST Measurement
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Statistical Analysis
Get Radiology Tree app to read full this article<
Results
Manual Measurement
Get Radiology Tree app to read full this article<
Global and Refined Registration
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Baseline and Follow-up Segmentation
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
RECIST Measurements
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Limitations
Get Radiology Tree app to read full this article<
Future Applications
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
References
1. Eisenhauer E.A., Therasse P., Bogaerts J., et. al.: New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: pp. 228-247.
2. Therasse P., Arbuck S.G., Eisenhauer E.A., et. al.: New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000; 92: pp. 205-216.
3. Jaffe T.A., Wickersham N.W., Sullivan D.C.: Quantitative imaging in oncology patients: part 1, radiology practice patterns at major U.S. cancer centers. AJR Am J Roentgenol 2010; 195: pp. 101-106.
4. Levy M.A., Rubin D.L.: Tool support to enable evaluation of the clinical response to treatment. AMIA Annu Symp Proc 2008 Nov 6;; pp. 399-403.
5. Hawkes D.J.: Algorithms for radiological image registration and their clinical application. J Anat 1998; 193: pp. 347-361.
6. Hutton B.F., Braun M., Thurfjell L., et. al.: Image registration: an essential tool for nuclear medicine. Eur J Nucl Med Mol Imaging 2002; 29: pp. 559-577.
7. Maintz J.B., Viergever M.A.: A survey of medical image registration. Med Image Anal 1998; 2: pp. 1-36.
8. Knops Z.F., Maintz J.B., Viergever M.A., et. al.: Normalized mutual information based registration using k-means clustering and shading correction. Med Image Anal 2006; 10: pp. 432-439.
9. Kuhnigk J.M., Dicken V., Bornemann L., et. al.: Morphological segmentation and partial volume analysis for volumetry of solid pulmonary lesions in thoracic CT scans. IEEE Trans Med Imaging 2006; 25: pp. 417-434.
10. Awad J., Owrangi A., Villemaire L., et. al.: Three-dimensional lung tumor segmentation from x-ray computed tomography using sparse field active models. Med Phys 2012; 39: pp. 851-865.
11. Xu T., Mandal M., Long R., et. al.: Gradient vector flow based active shape model for lung field segmentation in chest radiographs. Conf Proc IEEE Eng Med Biol Soc 2009; 2009: pp. 3561-3564.
12. Bauer S., Nolte L.P., Reyes M.: Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. Med Image Comput Comput Assist Interv 2011; 14: pp. 354-361.
13. Foo J.L., Miyano G., Lobe T., et. al.: Tumor segmentation from computed tomography image data using a probabilistic pixel selection approach. Comput Biol Med 2011; 41: pp. 56-65.
14. Kalkmann J., Ladd S.C., de Greiff A., et. al.: Suitability of semi-automated tumor response assessment of liver metastases using a dedicated software package. Rofo 2010; 182: pp. 581-588.
15. Sofka M., Stewart C.V.: Location registration and recognition (LRR) for serial analysis of nodules in lung CT scans. Med Image Anal 2010; 14: pp. 407-428.
16. Hong H., Lee J., Yim Y.: Automatic lung nodule matching on sequential CT images. Comput Biol Med 2008; 38: pp. 623-634.
17. Xu J., Greenspan H., Napel S., et. al.: Automated temporal tracking and segmentation of lymphoma on serial CT examinations. Med Phys 2011; 38: pp. 5879-5886.
18. Keil S., Behrendt F.F., Stanzel S., et. al.: RECIST and WHO criteria evaluation of cervical, thoracic and abdominal lymph nodes in patients with malignant lymphoma: manual versus semi-automated measurement on standard MDCT slices. Rofo 2009; 181: pp. 888-895.
19. Bland J.M., Altman D.G.: Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986; 1: pp. 307-310.
20. Pauls S., Kurschner C., Dharaiya E., et. al.: Comparison of manual and automated size measurements of lung metastases on MDCT images: potential influence on therapeutic decisions. Eur J Radiol 2008; 66: pp. 19-26.
21. Warr D., McKinney S., Tannock I.: Influence of measurement error on assessment of response to anticancer chemotherapy: proposal for new criteria of tumor response. J Clin Oncol 1984; 2: pp. 1040-1046.
22. Bornemann L., Kuhnigk J.M., Dicken V., et. al.: Informatics in radiology (infoRAD): new tools for computer assistance in thoracic CT part 2. Therapy monitoring of pulmonary metastases. Radiographics 2005; 25: pp. 841-848.
23. Schwartz L.H., Mazumdar M., Brown W., et. al.: Variability in response assessment in solid tumors: effect of number of lesions chosen for measurement. Clin Cancer Res 2003; 9: pp. 4318-4323.
24. Andreopoulou E., Andreopoulos D., Adamidis K., et. al.: Tumor volumetry as predictive and prognostic factor in the management of ovarian cancer. Anticancer Res 2002; 22: pp. 1903-1908.
25. Sohaib S.A., Turner B., Hanson J.A., et. al.: CT assessment of tumour response to treatment: comparison of linear, cross-sectional and volumetric measures of tumour size. Br J Radiol 2000; 73: pp. 1178-1184.
26. Yankelevitz D.F., Reeves A.P., Kostis W.J., et. al.: Small pulmonary nodules: volumetrically determined growth rates based on CT evaluation. Radiology 2000; 217: pp. 251-256.
27. Beer A.J., Wieder H.A., Lordick F., et. al.: Adenocarcinomas of esophagogastric junction: multi-detector row CT to evaluate early response to neoadjuvant chemotherapy. Radiology 2006; 239: pp. 472-480.
28. Lamata P., Lamata F., Sojar V., et. al.: Use of the Resection Map system as guidance during hepatectomy. Surg Endosc 2010; 24: pp. 2327-2337.
29. Conversano F., Franchini R., Demitri C., et. al.: Hepatic vessel segmentation for 3D planning of liver surgery experimental evaluation of a new fully automatic algorithm. Acad Radiol 2011; 18: pp. 461-470.
30. Koo C.W., Anand V., Girvin F., et. al.: Improved efficiency of CT interpretation using an automated lung nodule matching program. AJR Am J Roentgenol 2012; 199: pp. 91-95.
31. Folio LR, Sandouk A, Huang J, et al. Consistency and efficiency of computed tomography analysis in metastatic disease using a semi-automated lesion management application within PACS. AJR. 2013. In press
32. Huang J, Sandouk A, Folio LR. Accuracy of Volumes in a Phantom Serial CT Imaging. Presented at the Annual Meeting of the Radiological Society of North America, Chicago, IL. November 2012.