Rationale and Objectives
The discovery of germline genetic variants associated with breast cancer has engendered interest in risk stratification for improved, targeted detection and diagnosis. However, there has yet to be a comparison of the predictive ability of these genetic variants with mammography abnormality descriptors.
Materials and Methods
Our institutional review board-approved, Health Insurance Portability and Accountability Act-compliant study utilized a personalized medicine registry in which participants consented to provide a DNA sample and to participate in longitudinal follow-up. In our retrospective, age-matched, case-controlled study of 373 cases and 395 controls who underwent breast biopsy, we collected risk factors selected a priori based on the literature, including demographic variables based on the Gail model, common germline genetic variants, and diagnostic mammography findings according to Breast Imaging Reporting and Data System (BI-RADS). We developed predictive models using logistic regression to determine the predictive ability of (1) demographic variables, (2) 10 selected genetic variants, or (3) mammography BI-RADS features. We evaluated each model in turn by calculating a risk score for each patient using 10-fold cross-validation, used this risk estimate to construct Receiver Operator Characteristic Curve (ROC) curves, and compared the area under the ROC curve (AUC) of each using the DeLong method.
Results
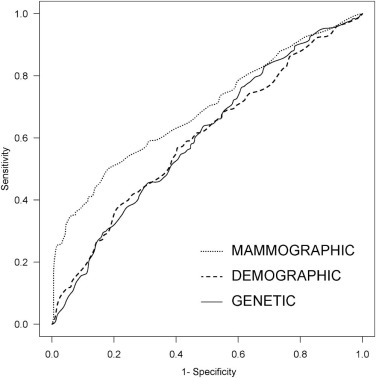
The performance of the regression model using demographic risk factors was not statistically different from the model using genetic variants ( P = 0.9). The model using mammography features (AUC = 0.689) was superior to both the demographic model (AUC = .598; P < 0.001) and the genetic model (AUC = .601; P < 0.001).
Conclusions
BI-RADS features exceeded the ability of demographic and 10 selected germline genetic variants to predict breast cancer in women recommended for biopsy.
Introduction
Over the last several decades, predictive variables have been discovered and incorporated into risk prediction models with the goal of personalizing breast cancer screening and diagnosis. One highly predictive source of information is abnormality level feature descriptors observed on mammography as described in the Breast Imaging Reporting and Data System (BI-RADS) . Other emerging sources are the ever-growing genome-wide association studies (GWAS) that identify genetic variants (single nucleotide polymorphisms [SNPs]). The SNPs discovered via recent GWAS are distinct from mutations in the BRCA1 and BRCA2 tumor suppressor genes . Although both are germline genetic risk factors (inherited from parental lineage), SNPs discovered in recent GWAS are single base pair DNA sequence variations conferring modest risk (low penetrance) but occurring commonly (high frequency) within the human population. Expansion of genetic risk prediction may depend on polygenic risk stratification, that is, weighing many high-frequency, low-penetrance SNPs at once . Early attempts to use such SNPs to predict breast cancer risk have demonstrated only modest improvements over conventional demographic risk factors, like those in the Gail model .
Breast cancer risk is determined by a combination of genetic and environmental factors. Intermediate phenotypes like imaging can capture and convey these interactions of these risk factors and provide biomarkers that can augment comprehensive risk prediction. Because demographic risk factors, genetic variants, and imaging features will all likely have some level of predictive value, determining which variables provide the best predictive power in any given setting becomes extremely important. Investing limited resources in collection of the best predictive variables will provide the most benefit. Prior literature evaluated risk prediction with genetics and breast density and one paper added BI-RADS assessment category . Despite the proven predictive ability of abnormality-level features described in the BI-RADS lexicon (e.g. mass and calcification descriptors as well as associated findings like architectural distortion), comparison to demographic or genetic risk has been limited. To estimate breast cancer risk in women recommended for breast biopsy, we compare the performance of predictive models using distinct data elements: demographic risk factors, germline genetic variants, or mammography abnormality features.
Materials and Methods
Subjects
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Epidemiological Risk Factors
Get Radiology Tree app to read full this article<
Genetic Variants
Get Radiology Tree app to read full this article<
Table 1
Common Genetic Variants Associated with Breast Cancer
SNPs Chromosome Gene High-risk Allele Low-risk Allele RS1045485 2q CASP8 G C RS13281615 8q Unknown G A RS13387042 2q Unknown A G RS2981582 10q FGFR2 T C RS3803662 16q TOX3 T C RS3817198 11p LSP1 C T RS889312 5q MAP3K1 C A RS10941679 5p Unknown G A RS999737 14q RAD51L1 C T RS11249433 1p Unknown C T
SNP, single nucleotide polymorphism.
Get Radiology Tree app to read full this article<
Mammography Features
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Statistical Analysis
Get Radiology Tree app to read full this article<
Table 2
Demographic Variables, Genetic Factors, and Mammographic Features in the Predictive Models \*
Variable Cases
( N = 373) Controls
( N = 395) OR 95% CI ¥P Value ¥ Age at menarche ≥14 105 (28.2%) 84 (21.2%) Referent 12–13 189 (50.7%) 224 (56.6%) 0.63 (0.33,1.21) 0.15 7–11 79 (21.1%) 88 (22.2%) 0.73 (0.29,1.83) 0.46 Number of biopsies 0 303 (81.2%) 337 (85.6%) Referent 1 60 (16.0%) 52 (13.1%) 1.35 (0.84,2.16) 0.21 ≥2 10 (2.6%) 6 (1.5%) 2.89 (0.95,8.75) 0.061 Number of pregnancies ≥6 52 (14.0%) 47 (12.0%) Referent 3–5 164 (43.9%) 175 (44.4%) 1.09 (0.63,1.89) 0.76 1–2 126 (33.8%) 129 (32.7%) 1.18 (0.66,2.13) 0.57 0 31 (8.3%) 43 (10.9%) 0.72 (0.34,1.51) 0.38 Number of first-degree relatives with breast cancer 0 268 (71.8%) 325 (82.2%) Referent 1 91 (24.4%) 57 (14.4%)2.05(1.34,3.15)0.0010 ≥2 14 (3.7%) 13 (3.2%) 1.68 (0.70,4.04) 0.24 Number of risk-conferring variant alleles 0–6 26 (6.9%) 58 (14.6%) Referent 7 or 8 97 (26.0%) 126 (31.9%)1.92(1.04,3.55)0.037 9 or 10 135 (36.1%) 126 (31.9%)2.69(1.46,4.95)0.0016 11 or 12 90 (24.1%) 73 (18.4%)2.74(1.41,5.30)0.0029 ≥13 25 (6.7%) 12 (3.0%)4.95(1.89,12.96)0.0012 Breast density Fatty 24 (6.5%) 26 (6.5%) Referent Scattered 30 (8.1%) 58 (14.6%) 0.46 (0.12,1.80) 0.24 Heterogeneous 293 (78.6%) 298 (75.4%) 0.99 (0.25,3.92) 0.98 Extremely dense 25 (6.8%) 14 (3.4%) 2.47 (0.27,22.27) 0.37 Mass margin Circumscribed 16 (4.2%) 36 (9.1%)0.42(0.21,0.85)0.016 Obscured 8 (2.1%) 14 (3.5%) 0.59 (0.22,1.58) 0.29 Ill defined 47 (12.6%) 48 (12.1%) 1.42 (0.87,2.31) 0.16 Spiculated 82 (21.9%) 4 (1.0%)27.56(9.64,78.78)<0.001 Suspicious microcalcification shape 63 (16.8%) 79 (20.0%) 1.49 (0.82,2.72) 0.19 Suspicious microcalcification distribution 79 (21.1%) 117 (29.6%)0.56(0.33,0.94)0.029 Architectural distortion 50 (13.4%) 21 (5.3%)2.32(1.25,4.28)0.0073
¥ Bold signifies statistically significant variables.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Assessment of Model Performance
Get Radiology Tree app to read full this article<
Institutional Review Board (IRB) Approvals
Get Radiology Tree app to read full this article<
Results
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 3
Frequency of Microcalcification Descriptors in Cases and Controls
Control Case Total Microcalcification shape Benign descriptor 33 (53%) 29 (47%) 62 Amorphous 10 (83%) 2 (17%) 12 Pleomorphic 70 (54%) 59 (46%) 129 Fine linear 0 (0%) 5 (100%) 5 Total 113 95 208 Microcalcification distribution Benign descriptor 11 (52%) 10 (48%) 21 Grouped 115 (62%) 71 (38%) 186 Segmental 0 (0%) 6 (100%) 6 Linear 18 (46%) 21 (54%) 39 Total 144 108 252
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Model Performance
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Funding
Get Radiology Tree app to read full this article<
Acknowledgments
Get Radiology Tree app to read full this article<
Appendix
Get Radiology Tree app to read full this article<
Table A1
Performance of MAMMOGRAPHIC Models Using Progressively More Recent Subsets of the Data
Time Frame_n_ AUC 1989–2010 768 .693 1993–2010 743 .701 1998–2010 662 .711
Get Radiology Tree app to read full this article<
References
1. Gail M.H., Brinton L.A., Byar D.P., et. al.: Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst 1989; 81: pp. 1879-1886.
2. Barlow W.E., White E., Ballard-Barbash R., et. al.: Prospective breast cancer risk prediction model for women undergoing screening mammography. J Natl Cancer Inst 2006; 98: pp. 1204-1214.
3. Pashayan N., Duffy S.W., Chowdhury S., et. al.: Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br J Cancer 2011; 104: pp. 1656-1663.
4. Baker J.A., Kornguth P.J., Lo J.Y., et. al.: Breast cancer: prediction with artificial neural network based on BI-RADS® standardized lexicon. Radiology 1995; 196: pp. 817-822.
5. Liberman L., Abramson A.F., Squires F.B., et. al.: The breast imaging reporting and data system: positive predictive value of mammographic features and final assessment categories. AJR Am J Roentgenol 1998; 171: pp. 35-40.
6. Burnside E.S., Rubin D.L., Fine J.P., et. al.: Bayesian network to predict breast cancer risk of mammographic microcalcifications and reduce number of benign biopsy results: initial experience. Radiology 2006; 240: pp. 666-673.
7. Chhatwal J., Alagoz O., Lindstrom M.J., et. al.: A logistic regression model based on the national mammography database format to aid breast cancer diagnosis. AJR Am J Roentgenol 2009; 192: pp. 1117-1127.
8. Timmers J.M., Verbeek A.L., IntHout J., et. al.: Breast cancer risk prediction model: a nomogram based on common mammographic screening findings. Eur Radiol 2013; 23: pp. 2413-2419.
9. Linger R.J., Kruk P.A.: BRCA1 16 years later: risk-associated BRCA1 mutations and their functional implications. FEBS J 2010; 277: pp. 3086-3096.
10. Pharoah P.D., Antoniou A.C., Easton D.F., et. al.: Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med 2008; 358: pp. 2796-2803.
11. Wacholder S., Hartge P., Prentice R., et. al.: Performance of common genetic variants in breast-cancer risk models. N Engl J Med 2010; 362: pp. 986-993.
12. Gail M.H.: Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst 2008; 100: pp. 1037-1041.
13. Gail M.H.: Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst 2009; 101: pp. 959-963.
14. Devilee P., Rookus M.A.: A tiny step closer to personalized risk prediction for breast cancer. N Engl J Med 2010; 362: pp. 1043-1045.
15. Tamimi R.M., Cox D., Kraft P., et. al.: Breast cancer susceptibility loci and mammographic density. Breast Cancer Res 2008; 10: pp. R66.
16. Darabi H., Czene K., Zhao W., et. al.: Breast cancer risk prediction and individualised screening based on common genetic variation and breast density measurement. Breast Cancer Res 2012; 14: pp. R25.
17. Armstrong K., Handorf E.A., Chen J., et. al.: Breast cancer risk prediction and mammography biopsy decisions: a model-based study. Am J Prev Med 2013; 44: pp. 15-22.
18. McCarty C.A., Wilke R.A., Giampietro P.F., et. al.: Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Med 2005; 2: pp. 49-79.
19. Wu Y., Alagoz O., Ayvaci M.U., et. al.: A comprehensive methodology for determining the most informative mammographic features. J Digit Imaging 2013; 26: pp. 941-947.
20. Kobayashi S., Sugiura H., Ando Y., et. al.: Reproductive history and breast cancer risk. Breast Cancer 2012; 19: pp. 302-308.
21. McCarty C.A., Chisholm R.L., Chute C.G., et. al.: The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4: pp. 13.
22. Easton D.F., Pooley K.A., Dunning A.M., et. al.: Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007; 447: pp. 1087-1093.
23. Hunter D.J., Kraft P., Jacobs K.B., et. al.: A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 2007; 39: pp. 870-874.
24. American College of Radiology : 4th ed.2003.American College of RadiologyReston VA
25. American College of Radiology : 3rd ed.1998.American College of RadiologyReston VA
26. Houssam N., Ryan W., Elizabeth B., et. al.: Information Extraction for Clinical Data Mining: A Mammography Case Study. In: Proceedings of the 2009 IEEE International Conference on Data Mining Workshops: IEEE Computer Society2009.
27. Percha B., Nassif H., Lipson J., et. al.: Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc 2012; 19: pp. 913-916.
28. Swets J.A.: Measuring the accuracy of diagnostic systems. Science 1988; 240: pp. 1285-1293.
29. Burnside E.S., Sickles E.A., Bassett L.W., et. al.: The ACR BI-RADS experience: learning from history. J Am Coll Radiol 2009; 6: pp. 851-860.
30. Rubin D.B.: Multiple imputation for nonresponse in surveys.1987.John Wiley & SonsNew York
31. van Buuren S.: Flexible imputation of missing data.CRC/Chapman & Hall Interdisciplinary Statistics Series2012.Chapman and Hall/CRCBoca Raton, FL
32. van Buuren S., Oudshoorn C.G.M.: Multivariate Imputation by Chained Equations MICE V1.0 User’s manual. Leiden, The Netherlands: TNO Prevention and Health; June2000. Report No.: 90-6743-677-1
33. van Buuren S., Groothuis-Oudshoorn K.: mice: Multivariate Imputation by Chained Equations in R. J Stat Softw 2011; 45: pp. 1-67.
34. Van Buuren S., Oudshoorn C.G.M.: Multivariate imputation by chained equations: MICE V1.0 user’s manual.2000.TNO Prevention and HealthLeiden
35. DeLong E.R., DeLong D.M., Clarke-Pearson D.L.: Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: pp. 837-845.
36. R Core Team : 2013.R Foundation for Statistical ComputingVienna, Austria [3.0.1]
37. Tice J.A., Cummings S.R., Smith-Bindman R., et. al.: Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Ann Intern Med 2008; 148: pp. 337-347.
38. Berg W.A., Arnoldus C.L., Teferra E., et. al.: Biopsy of amorphous breast calcifications: pathologic outcome and yield at stereotactic biopsy. Radiology 2001; 221: pp. 495-503.
39. Burnside E.S., Ochsner J.E., Fowler K.J., et. al.: Use of microcalcification descriptors in BI-RADS 4th edition to stratify risk of malignancy. Radiology 2007; 242: pp. 388-395.
40. Michailidou K., Hall P., Gonzalez-Neira A., et. al.: Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 2013; 45: pp. 353-361. 361e1-2