One in six men will develop prostate cancer in his lifetime. Early detection and accurate diagnosis of the disease can improve cancer survival and reduce treatment costs. Recently, imaging of prostate cancer has greatly advanced since the introduction of multiparametric magnetic resonance imaging (mp-MRI). Mp-MRI consists of T2-weighted sequences combined with functional sequences including dynamic contrast-enhanced MRI, diffusion-weighted MRI, and magnetic resonance spectroscopy imaging. Because of the big data and variations in imaging sequences, detection can be affected by multiple factors such as observer variability and visibility and complexity of the lesions. To improve quantitative assessment of the disease, various computer-aided detection systems have been designed to help radiologists in their clinical practice. This review paper presents an overview of literatures on computer-aided detection of prostate cancer with mp-MRI, which include the technology and its applications. The aim of the survey is threefold: an introduction for those new to the field, an overview for those working in the field, and a reference for those searching for literature on a specific application.
Introduction
Prostate cancer (PCa) is currently the most common cancer in men and the second leading cause of cancer-related deaths among men in the United States . In 2015, it is estimated that the number of estimated new cases and deaths will be 220,800 and 27,540, respectively, accounting for 26.0% of new cancer cases and 8.8% of cancer deaths for American men .
The prostate is subdivided into the base, mid-gland, and apex from superior to inferior. The prostate also has four anatomic zones: the transition zone (TZ), which contains 5% of the glandular tissue and accounts for around 25% of PCa; the central zone, which contains 20% of the glandular tissue and accounts for around 5% of PCa; the peripheral zone (PZ), which contains 70–80% of the glandular tissue and accounts for about 70% of PCa; and the non-glandular anterior fibromuscular stroma. Accurate localization of PCa within the TZ or the PZ is extremely important as TZ PCa is associated with favorable pathologic features and better recurrence-free survival .
At present, the clinical standard for definitive diagnosis of PCa is transrectal ultrasound (TRUS)-guided sextant or systematic biopsy. The prostate-specific antigen (PSA) blood test and digital rectal examination (DRE) results are considered to identify patients who need biopsy. The actual impact of magnetic resonance imaging (MRI) for PCa management is through guided biopsies and improved cancer diagnosis and staging yield. In recent years, MRI-targeted prostate biopsies have been showing better disease localization and more accurate sampling than conventional TRUS-guided biopsy in various studies . MRI-based computer-assisted sophisticated imaging for individual patients would offer such a significant role in defining an optimal targeted biopsy and interventional approach. Several approaches have been explored to improve the accuracy of image-guided targeted prostate biopsy, including in-bore MRI-guided, cognitive fusion, and MRI/TRUS fusion-guided biopsy .
MRI provides excellent soft-tissue contrast and has become an imaging modality of choice for localization of prostate tumors. Multiparametric MRI (mp-MRI) includes high-resolution T2-weighted (T2W) MRI, diffusion-weighted imaging (DWI), dynamic contrast-enhanced imaging (DCE-MR), and MR spectroscopy (MRS). The mp-MRI has proven to be an effective technique to localize high-risk PCa . The combined use of anatomic and functional information provided by the multiparametric approach increases the accuracy of MRI in detecting and staging PCa . It can also help guide biopsies to achieve a higher tumor detection rate and better reflect the true Gleason grade. The European Society of Urogenital Radiology in 2012 established the Prostate Imaging Reporting and Data System (PI-RADS) scoring system for mp-MRI of the prostate . The MR PI-RADS aims to enable consistent interpretation, communication, and reporting of prostate mp-MRI findings . A joint steering committee formed by the American College of Radiology, European Society of Urogenital Radiology, and the AdMeTech Foundation has recently announced an updated version of the proposals of PI-RADS Version 2 . Prostate mp-MRI at 3 T had been recommended in PI-RADS Version 2. Generally, computer-aided detection (CAD) systems are classified into two categories: CAD and computer-aided diagnosis (CADx) systems. Currently, most CAD systems in prostate MRI focus on local suspicious lesions and discrimination between benign and malignant lesions; most of them are CADx systems. As the combination of various MR images creates large amounts of data, supportive techniques or tools, such as CADx, are needed to make a clinical decision in a fast, effective, and reliable way.
In the past 10 years, computer-aided techniques have developed rapidly. Automated CAD and diagnosis may help improve diagnostic accuracy of PCa, and reduce interpretation variation between and within observers . PCa diagnosis requires an experienced radiologist to read prostate MRI, and such expertise is not widely available. Addition of CADx may significantly improve the performance of less-experienced observers in PCa diagnosis. When less-experienced observers used CADx, they had a similar performance as those experienced observers in distinguishing benign from malignant lesions . In a more recent study, the use of CAD can also improve prostate mp-MRI study interpretation in experienced readers . For cases in which radiologists are less confident, they can get higher performance by using the computer output. A recent study showed that a pattern recognition system enables radiologists to have a lower variability in diagnosis, decreases false-negative rates, and reduces the time to recognize and delineate structures in the prostate . The benefit of CADx also includes guiding biopsy using cancer location information from MRI . Therefore, along with rapid development of MR technique, CADx of PCa has become an active field of research in the last 5 years.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
MR Image Acquisitions
Get Radiology Tree app to read full this article<
T2WI and T2 Mapping
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
DCE-MRI
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Diffusion-weighted MRI
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
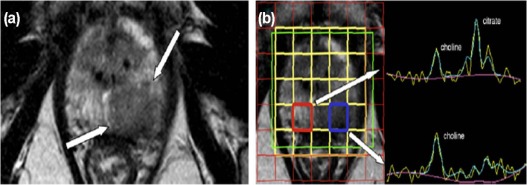
MRS
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Other Imaging Methods
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
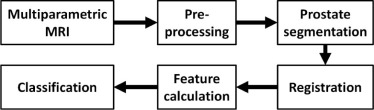
MR Image Quantification Methods
General Framework
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Preprocessing
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
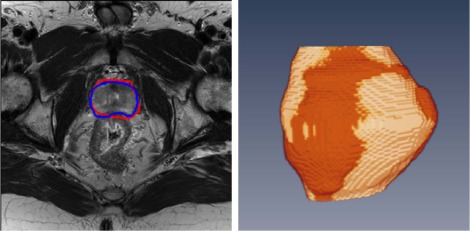
Segmentation
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
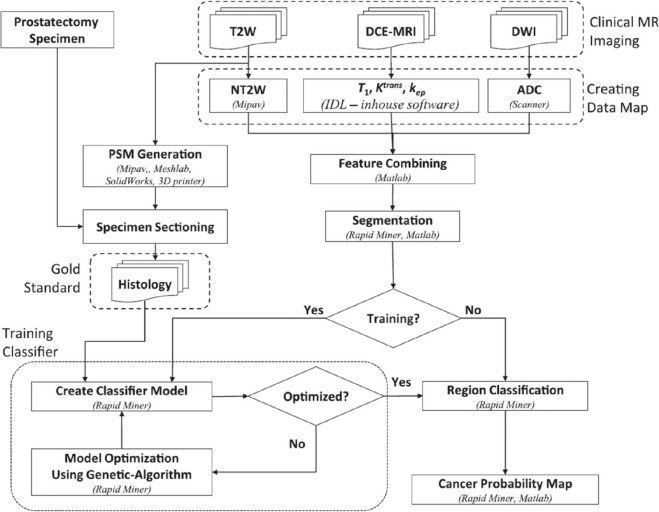
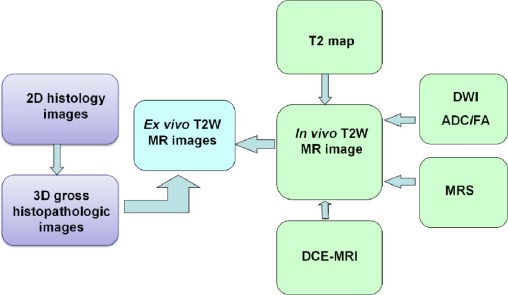
Registration
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Feature Extraction
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Classification
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Validation
Get Radiology Tree app to read full this article<
Table 1
Validation of CADx Systems
Reference Ground Truth on the Histology Candidate on MR Image Image Registration Chan et al. Biopsy MO NA Puech et al. Needle biopsy or prostatectomy MO NA Tiwari et al. Biopsy Sextant location determined by radiologist NA Vos et al. WMHS + MO MO 3D rendering mode Viswanath et al. WMHS + MO MANTRA Multimodal image registration Viswanath et al. WMHS MANTRA Multimodal image registration Vos et al. WMHS Not specified Not specified Liu et al. WMHS + MO MO + ex vivo MRI Manual Tiwari et al. WMHS + sextant boundaries A joint review session of trial imagers and pathologists NA Artan et al. WMHS + MO Tumor location is transferred to the in vivo MRI from histologic images + ex vivo MRI NA Vos et al. WMHS + MO MO Mutual information registration Viswanath et al. WMHS + MO Registration from histologic images MACMI Lopes et al. WMHS + drawn by urologists Drawn by urologists Manual correspondence Liu and Yetik WMHS + MO MO + ex vivo MRI Manual registration Sung et al. Radical prostatectomy + MO The radiologist matched the pathologic slices with corresponding MRI NA Tiwari et al. WMHS MO + ex vivo MRI Manual registration Viswanath et al. WMHS + MO Registration from histologic images Multimodal elastic registration Vos et al. Needle biopsy Combining the findings with, histopathology of MR-guided samples by radiologist. NA Niaf et al. WMHS + MO MO Manual registration Artan and Yetik WMHS + MO MO + ex vivo MRI Manual registration Shah et al. WMHS + MO Not specified PSM Matulewicz et al. WMHS + MO MO Manual registration Hambrock et al. WMHS + MO MO Manual registration Tiwari et al. WMHS + MO MO Manual registration Peng et al. WMHS MO Manual registration Ginsburg et al. WMHS + MO Registration from histologic images Nonlinear registration Stember et al. Needle biopsy Not specified NA Niaf et al. Prostatectomy + MO MO Manual registration Garcia Molina et al. Prostatectomy + MO MO Manual registration Litjens et al. Needle biopsy Not specified NA Kwak et al. Needle biopsy Determined by radiologists NA Zhao et al. Biopsy MO NA
3D, three-dimensional; CADx, computer-aided diagnosis; MACMI, multi-attribute, higher order mutual information based elastic registration scheme; MANTRA, multi-attribute, non-initializing, texture reconstruction based active shape model (ASM); MO, manual outlined regions of lesions; MR, magnetic resonance; NA, no registration was used; PSM, patient-specific molds; WMHS, whole-mount histologic sections.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Clinical Applications
Diagnosis
Get Radiology Tree app to read full this article<
Table 2
Summary of Representative Studies in the Literature
Reference Modality Validation Region Classifier Data Size Performance Chan et al. T2WI, ADC, T2 Biopsy PZ SVM, FLD 15 FLD, AUC = 0.839; SVM, AUC = 0.761 Puech et al. DCE Prostatectomy PZ and TZ Software titled “ProCAD” 100 PZ, Se/Sp = 100/49%; TZ, Se/Sp = 100/40% Tiwari et al. MRS Biopsy WP Spectral clustering 14 Se = 77.8%, FP = 28.92%, and FN = 20.88% Vos et al. DCE WMHS PZ SVM 34 AUC = 0.83 Viswanath et al. DCE WMHS WP LLE and consensus clustering 6 Se = 60.72%, Sp = 83.24% Viswanath et al. T2WI, DCE WMHS WP Random forest 6 AUC = 0.815 Vos et al. DCE WMHS PZ SVM 38 AUC = 0.80 Liu et al. T2W, T2, ADC, DCE WMHS PZ Fuzzy MRF model 11 Se = 89.58%, Sp = 87.50% Tiwari et al. MRS Prostatectomy WP NLDR 18 Se = 89.33%, Sp = 79.79% Artan et al. T2, ADC, DCE Biopsy PZ Cost-sensitive CRF 21 AUC = 0.79 Vos et al. T2WI, DCE WMHS PZ SVM 29 AUC = 0.89 Viswanath et al. T2W, DWI, DCE WMHS WP EMPrAvISE 12 AUC = 0.77 Lopes et al. T2WI WMHS WP SVM, AdaBoost 17 SVM, Se/Sp = 83/91%; AdaBoost, Se/Sp = 85/93% Liu and Yetik T2W, DWI, DCE WMHS WP SVM 20 AUC = 0.89 Sung et al. DCE Prostatectomy PZ and TZ SVM 42 PZ, Se/Sp = 89/89%; TZ, Se/Sp = 91/64% Tiwari et al. T2WI, MRS WMHS WP Random forest 36 AUC = 0.89 Viswanath et al. T2WI WMHS PZ and CG QDA 22 CG, AUC = 0.86; PZ, AUC = 0.73 Vos et al. T1, T2, ADC, DCE Biopsy WP LDA 200 Se = 0.74, at an FP level of 5 per patient Niaf et al. T2W, DWI, DCE WMHS PZ SVM 30 AUC = 0.89 Artan and Yetik T2, ADC, T1-PC WMHS WP SVM 15 Se = 76%, Sp = 86% Shah et al. T2WI, ADC, DCE WMHS PZ SVM 31 f-measure = 89% Matulewicz et al. MRS WMHS WP ANN 18 AUC = 0.968 Hambrock et al. T2WI, DWI, DCE Prostatectomy PZ and TZ In-house–developed CAD system 34 Experienced observers, AUC = 0.91 Tiwari et al. T2WI, MRS WMHS WP SeSMiK-GE 29 AUC = 0.89 Peng et al. T2WI, ADC, DCE Prostatectomy WP LDA 48 AUC = 0.95 Ginsburg et al. T2WI, DWI, DCE WMHS PZ and CG PCA-VIP 108 CG, AUC = 0.85; PZ, AUC = 0.79 Stember et al. T2WI, ADC Biopsy TZ Naive Bayes classifier 18 Predicted TZ tumor in all test patients Niaf et al. T2WI, DWI, DCE Prostatectomy WP P-SVM 48 AUC = 0.889 Garcia Molina et al. T2WI, ADC, DCE Prostatectomy PZ Incremental learning ensemble SVM 12 Se = 84.4%,Sp = 78.0% Litjens et al. T2WI, DWI, DCE, PDWI Biopsy WP Random forest 347 AUC = 0.889 Kwak et al. T2WI, DWI Biopsy PZ and TZ SVM 244 AUC of 0.89 Zhao et al. T2WI Biopsy/follow-up PZ and CG ANN 71 CG, AUC = 0.821; PZ, AUC = 0.849
ADC, apparent diffusion coefficient; ANN, artificial neural network; AUC, area under a receiver operating characteristic curve; CAD, computer-aided detection; CG, central gland; CRF, conditional random fields; DCE, dynamic contrast-enhanced; EMPrAvISE, Enhanced Multi-Protocol Analysis via Intelligent Supervised Embedding; FLD, Fisher linear discriminant; FN, false negative; FP, false positive; LDA, linear discriminant analysis; LLE, locally linear embedding; MRS, magnetic resonance spectroscopy; NLDR, nonlinear dimensionality reduction; PCA, principal component analysis; PCA-VIP, variable importance on projection measure for PCA; P-SVM, probabilistic SVM; PZ, peripheral zone; QDA, quadratic discriminant analysis; Se, sensitivity; SeSMiK-GE, Semi Supervised Multi Kernel Graph Embedding; Sp, specificity; SVM, support vector machine; T1-PC, principal component of T1-weighted dynamic series; T2WI, T2-weighted imaging; TZ, transition zone; WMHS, whole-mount histologic sections; WP, whole prostate.
Get Radiology Tree app to read full this article<
Aggressiveness
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Biopsy Guidance
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Treatment Planning and Therapeutic Response Assessment
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion and Future Directions
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusion
Get Radiology Tree app to read full this article<
Acknowledgments
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Siegel R.L., Miller K.D., Jemal A.: Cancer statistics, 2015. CA Cancer J Clin 2015; 65: pp. 5-29.
2. Lee J.J., Thomas I.C., Nolley R., et. al.: Biologic differences between peripheral and transition zone prostate cancer. Prostate 2015; 75: pp. 183-190.
3. Siddiqui M.M., Rais-Bahrami S., Turkbey B., et. al.: Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015; 313: pp. 390-397.
4. Pokorny M.R., de Rooij M., Duncan E., et. al.: Prospective study of diagnostic accuracy comparing prostate cancer detection by transrectal ultrasound-guided biopsy versus magnetic resonance (MR) imaging with subsequent MR-guided biopsy in men without previous prostate biopsies. Eur Urol 2014; 66: pp. 22-29.
5. Schoots I.G., Roobol M.J., Nieboer D., et. al.: Magnetic resonance imaging-targeted biopsy may enhance the diagnostic accuracy of significant prostate cancer detection compared to standard transrectal ultrasound-guided biopsy: a systematic review and meta-analysis. Eur Urol 2015; 68: pp. 438-450.
6. Zhang J., Xiu J., Dong Y., et. al.: Magnetic resonance imaging-directed biopsy improves the prediction of prostate cancer aggressiveness compared with a 12-core transrectal ultrasound-guided prostate biopsy. Mol Med Rep 2014; 9: pp. 1989-1997.
7. Brown A.M., Elbuluk O., Mertan F., et. al.: Recent advances in image-guided targeted prostate biopsy. Abdom Imaging 2015; 40: pp. 1788-1799.
8. Puech P., Rouviere O., Renard-Penna R., et. al.: Prostate cancer diagnosis: multiparametric MR-targeted biopsy with cognitive and transrectal US-MR fusion guidance versus systematic biopsy—prospective multicenter study. Radiology 2013; 268: pp. 461-469.
9. Lawrence E.M., Tang S.Y., Barrett T., et. al.: Prostate cancer: performance characteristics of combined T(2)W and DW-MRI scoring in the setting of template transperineal re-biopsy using MR-TRUS fusion. Eur Radiol 2014; 24: pp. 1497-1505.
10. Barentsz J.O., Richenberg J., Clements R., et. al.: ESUR prostate MR guidelines. Eur Radiol 2012; 2012: pp. 746-757.
11. Dickinson L., Ahmed H.U., Allen C., et. al.: Magnetic resonance imaging for the detection, localisation, and characterisation of prostate cancer: recommendations from a European consensus meeting. Eur Urol 2011; 59: pp. 477-494.
12. Weinreb J.C., Barentsz J.O., Choyke P.L., et. al.: PI-RADS Prostate Imaging-Reporting and Data System: 2015, version 2. Eur Urol 2016; 69: pp. 16-40.
13. Hambrock T., Vos P.C., Hulsbergen-van de Kaa C.A., et. al.: Prostate cancer: computer-aided diagnosis with multiparametric 3-T MR imaging—effect on observer performance. Radiology 2013; 266: pp. 521-530.
14. Vos P.C., Barentsz J.O., Karssemeijer N., et. al.: Automatic computer-aided detection of prostate cancer based on multiparametric magnetic resonance image analysis. Phys Med Biol 2012; 57: pp. 1527-1542.
15. Niaf E., Lartizien C., Bratan F., et. al.: Prostate focal peripheral zone lesions: characterization at multiparametric MR imaging—influence of a computer-aided diagnosis system. Radiology 2014; 271: pp. 761-769.
16. Garcia Molina J.F., Zheng L., Sertdemir M., et. al.: Incremental learning with SVM for multimodal classification of prostatic adenocarcinoma. PLoS ONE 2014; 9: pp. e93600.
17. Shah V., Turkbey B., Mani H., et. al.: Decision support system for localizing prostate cancer based on multiparametric magnetic resonance imaging. Med Phys 2012; 39: pp. 4093-4103.
18. Niaf E., Rouviere O., Mege-Lechevallier F., et. al.: Computer-aided diagnosis of prostate cancer in the peripheral zone using multiparametric MRI. Phys Med Biol 2012; 57: pp. 3833-3851.
19. Akin O., Sala E., Moskowitz C.S., et. al.: Transition zone prostate cancers: features, detection, localization, and staging at endorectal MR imaging. Radiology 2006; 239: pp. 784-792.
20. Li H., Sugimura K., Kaji Y., et. al.: Conventional MRI capabilities in the diagnosis of prostate cancer in the transition zone. AJR Am J Roentgenol 2006; 186: pp. 729-742.
21. Cornud F., Rouanne M., Beuvon F., et. al.: Endorectal 3D T2-weighted 1 mm-slice thickness MRI for prostate cancer staging at 1.5Tesla: should we reconsider the indirects signs of extracapsular extension according to the D’Amico tumor risk criteria?. Eur J Radiol 2012; 81: pp. e591-e597.
22. Viswanath S.E., Bloch N.B., Chappelow J.C., et. al.: Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 Tesla endorectal, in vivo T2-weighted MR imagery. J Magn Reson Imaging 2012; 36: pp. 213-224.
23. Hoeks C.M., Hambrock T., Yakar D., et. al.: Transition zone prostate cancer: detection and localization with 3-T multiparametric MR imaging. Radiology 2013; 266: pp. 207-217.
24. Liu W., Turkbey B., Senegas J., et. al.: Accelerated T2 mapping for characterization of prostate cancer. Magn Reson Med 2011; 65: pp. 1400-1406.
25. Yamauchi F.I., Penzkofer T., Fedorov A., et. al.: Prostate cancer discrimination in the peripheral zone with a reduced field-of-view T(2)-mapping MRI sequence. Magn Reson Imaging 2015; 33: pp. 525-530.
26. Liu X., Yetik I.S.: Automated prostate cancer localization without the need for peripheral zone extraction using multiparametric MRI. Med Phys 2011; 38: pp. 2986-2994.
27. Peng Y., Jiang Y., Yang C., et. al.: Quantitative analysis of multiparametric prostate MR images: differentiation between prostate cancer and normal tissue and correlation with Gleason score—a computer-aided diagnosis development study. Radiology 2013; 267: pp. 787-796.
28. Nowak J., Malzahn U., Baur A.D., et. al.: The value of ADC, T2 signal intensity, and a combination of both parameters to assess Gleason score and primary Gleason grades in patients with known prostate cancer. Acta Radiol 2016; 57: pp. 107-114.
29. Hoang Dinh A., Souchon R., Melodelima C., et. al.: Characterization of prostate cancer using T2 mapping at 3T: a multi-scanner study. Diagn Interv Imaging 2015; 96: pp. 365-372.
30. Durmus T., Baur A., Hamm B.: Multiparametric magnetic resonance imaging in the detection of prostate cancer. Aktuelle Urol 2014; 45: pp. 119-126.
31. Ewing J.R., Bagher-Ebadian H.: Model selection in measures of vascular parameters using dynamic contrast-enhanced MRI: experimental and clinical applications. NMR Biomed 2013; 26: pp. 1028-1041.
32. Tofts P.S., Brix G., Buckley D.L., et. al.: Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 1999; 10: pp. 223-232.
33. Fedorov A., Fluckiger J., Ayers G.D., et. al.: A comparison of two methods for estimating DCE-MRI parameters via individual and cohort based AIFs in prostate cancer: a step towards practical implementation. Magn Reson Imaging 2014; 32: pp. 321-329.
34. Gliozzi A.S., Mazzetti S., Delsanto P.P., et. al.: Phenomenological universalities: a novel tool for the analysis of dynamic contrast enhancement in magnetic resonance imaging. Phys Med Biol 2011; 56: pp. 573-586.
35. Mazzetti S., Gliozzi A.S., Bracco C., et. al.: Comparison between PUN and Tofts models in the quantification of dynamic contrast-enhanced MR imaging. Phys Med Biol 2012; 57: pp. 8443-8453.
36. Vos P.C., Hambrock T., Hulsbergen-van de Kaa C.A., et. al.: Computerized analysis of prostate lesions in the peripheral zone using dynamic contrast enhanced MRI. Med Phys 2008; 35: pp. 888-899.
37. Vos P.C., Hambrock T., Barenstz J.O., et. al.: Automated calibration for computerized analysis of prostate lesions using pharmacokinetic magnetic resonance images. Med Image Comput Comput Assist Interv 2009; 12: pp. 836-843.
38. Puech P., Betrouni N., Makni N., et. al.: Computer-assisted diagnosis of prostate cancer using DCE-MRI data: design, implementation and preliminary results. Int J Comput Assist Radiol Surg 2009; 4: pp. 1-10.
39. Puech P., Betrouni N., Viard R., et. al.: Prostate cancer computer-assisted diagnosis software using dynamic contrast-enhanced MRI. Conf Proc IEEE Eng Med Biol Soc 2007; 2007: pp. 5567-5570.
40. Padhani A.R., Gapinski C.J., Macvicar D.A., et. al.: Dynamic contrast enhanced MRI of prostate cancer: correlation with morphology and tumour stage, histological grade and PSA. Clin Radiol 2000; 55: pp. 99-109.
41. Dikaios N., Alkalbani J., Abd-Alazeez M., et. al.: Zone-specific logistic regression models improve classification of prostate cancer on multi-parametric MRI. Eur Radiol 2015; 25: pp. 2727-2737.
42. Yacoub J.H., Oto A., Miller F.H.: MR imaging of the prostate. Radiol Clin North Am 2014; 52: pp. 811-837.
43. Toivonen J., Merisaari H., Pesola M., et. al.: Mathematical models for diffusion-weighted imaging of prostate cancer using b values up to 2000 s/mm(2): correlation with Gleason score and repeatability of region of interest analysis. Magn Reson Med 2015; 74: pp. 1116-1124.
44. Boesen L., Chabanova E., Logager V., et. al.: Apparent diffusion coefficient ratio correlates significantly with prostate cancer Gleason score at final pathology. J Magn Reson Imaging 2015; 42: pp. 446-453.
45. Mazaheri Y., Shukla-Dave A., Hricak H., et. al.: Prostate cancer: identification with combined diffusion-weighted MR imaging and 3D 1H MR spectroscopic imaging—correlation with pathologic findings. Radiology 2008; 246: pp. 480-488.
46. Peng Y., Jiang Y., Antic T., et. al.: Validation of quantitative analysis of multiparametric prostate MR images for prostate cancer detection and aggressiveness assessment: a cross-imager study. Radiology 2014; 271: pp. 461-471.
47. Kitajima K., Takahashi S., Ueno Y., et. al.: Clinical utility of apparent diffusion coefficient values obtained using high b-value when diagnosing prostate cancer using 3 tesla MRI: comparison between ultra-high b-value (2000 s/mm(2)) and standard high b-value (1000 s/mm(2)). J Magn Reson Imaging 2012; 36: pp. 198-205.
48. Kim C.K., Park B.K., Kim B.: High-b-value diffusion-weighted imaging at 3 T to detect prostate cancer: comparisons between b values of 1000 and 2000 s/mm 2 . AJR Am J Roentgenol 2010; 194: pp. W33-W37.
49. Wetter A., Nensa F., Lipponer C., et. al.: High and ultra-high b-value diffusion-weighted imaging in prostate cancer: a quantitative analysis. Acta Radiol 2015; 56: pp. 1009-1015.
50. Wang X., Qian Y., Liu B., et. al.: High-b-value diffusion-weighted MRI for the detection of prostate cancer at 3 T. Clin Radiol 2014; 69: pp. 1165-1170.
51. Stember J.N., Deng F.M., Taneja S.S., et. al.: Pilot study of a novel tool for input-free automated identification of transition zone prostate tumors using T2- and diffusion-weighted signal and textural features. J Magn Reson Imaging 2014; 40: pp. 301-305.
52. Kwak J.T., Xu S., Wood B.J., et. al.: Automated prostate cancer detection using T2-weighted and high-b-value diffusion-weighted magnetic resonance imaging. Med Phys 2015; 42: pp. 2368-2378.
53. Neto J.A., Parente D.B.: Multiparametric magnetic resonance imaging of the prostate. Magn Reson Imaging Clin N Am 2013; 21: pp. 409-426.
54. Tiwari P., Madabhushi A., Rosen M.: A hierarchical unsupervised spectral clustering scheme for detection of prostate cancer from magnetic resonance spectroscopy (MRS). Med Image Comput Comput Assist Interv 2007; 10: pp. 278-286.
55. Tiwari P., Rosen M., Madabhushi A.: A hierarchical spectral clustering and nonlinear dimensionality reduction scheme for detection of prostate cancer from magnetic resonance spectroscopy (MRS). Med Phys 2009; 36: pp. 3927-3939.
56. Tiwari P., Viswanath S., Kurhanewicz J., et. al.: Multimodal wavelet embedding representation for data combination (MaWERiC): integrating magnetic resonance imaging and spectroscopy for prostate cancer detection. NMR Biomed 2012; 25: pp. 607-619.
57. Matulewicz L., Jansen J.F., Bokacheva L., et. al.: Anatomic segmentation improves prostate cancer detection with artificial neural networks analysis of 1H magnetic resonance spectroscopic imaging. J Magn Reson Imaging 2014; 40: pp. 1414-1421.
58. Panebianco V., Barchetti F., Sciarra A., et. al.: In vivo 3D neuroanatomical evaluation of periprostatic nerve plexus with 3T-MR diffusion tensor imaging. Eur J Radiol 2013; 82: pp. 1677-1682.
59. Sinha S., Sinha U.: In vivo diffusion tensor imaging of the human prostate. Magn Reson Med 2004; 52: pp. 530-537.
60. Xu J., Humphrey P.A., Kibel A.S., et. al.: Magnetic resonance diffusion characteristics of histologically defined prostate cancer in humans. Magn Reson Med 2009; 61: pp. 842-850.
61. Takayama Y., Kishimoto R., Hanaoka S., et. al.: ADC value and diffusion tensor imaging of prostate cancer: changes in carbon-ion radiotherapy. J Magn Reson Imaging 2008; 27: pp. 1331-1335.
62. Litjens G., Debats O., Barentsz J., et. al.: Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging 2014; 33: pp. 1083-1092.
63. Suo S., Chen X., Wu L., et. al.: Non-Gaussian water diffusion kurtosis imaging of prostate cancer. Magn Reson Imaging 2014; 32: pp. 421-427.
64. Roethke M.C., Kuder T.A., Kuru T.H., et. al.: Evaluation of diffusion kurtosis imaging versus standard diffusion imaging for detection and grading of peripheral zone prostate cancer. Invest Radiol 2015; 50: pp. 483-489.
65. Suga M., Aga T., Minato K.: Development of a magnetic resonance elastic microscope system. Conf Proc IEEE Eng Med Biol Soc 2004; 2: pp. 1025-1027.
66. Venkatesh S.K., Yin M., Glockner J.F., et. al.: MR elastography of liver tumors: preliminary results. AJR Am J Roentgenol 2008; 190: pp. 1534-1540.
67. Hamhaber U., Klatt D., Papazoglou S., et. al.: In vivo magnetic resonance elastography of human brain at 7 T and 1.5 T. J Magn Reson Imaging 2010; 32: pp. 577-583.
68. Sinkus R., Tanter M., Xydeas T., et. al.: Viscoelastic shear properties of in vivo breast lesions measured by MR elastography. Magn Reson Imaging 2005; 23: pp. 159-165.
69. McKnight A.L., Kugel J.L., Rossman P.J., et. al.: MR elastography of breast cancer: preliminary results. AJR Am J Roentgenol 2002; 178: pp. 1411-1417.
70. Sinkus R., Lorenzen J., Schrader D., et. al.: High-resolution tensor MR elastography for breast tumour detection. Phys Med Biol 2000; 45: pp. 1649-1664.
71. Arani A., Da Rosa M., Ramsay E., et. al.: Incorporating endorectal MR elastography into multi-parametric MRI for prostate cancer imaging: initial feasibility in volunteers. J Magn Reson Imaging 2013; 38: pp. 1251-1260.
72. Li S., Chen M., Wang W., et. al.: A feasibility study of MR elastography in the diagnosis of prostate cancer at 3.0 T. Acta Radiol 2011; 52: pp. 354-358.
73. Arani A., Plewes D., Krieger A., et. al.: The feasibility of endorectal MR elastography for prostate cancer localization. Magn Reson Med 2011; 66: pp. 1649-1657.
74. Fennessy F.M., Fedorov A., Gupta S.N., et. al.: Practical considerations in T1 mapping of prostate for dynamic contrast enhancement pharmacokinetic analyses. Magn Reson Imaging 2012; 30: pp. 1224-1233.
75. Fortuin A.S., Smeenk R.J., Meijer H.J., et. al.: Lymphotropic nanoparticle-enhanced MRI in prostate cancer: value and therapeutic potential. Curr Urol Rep 2014; 15: pp. 389.
76. Thoeny H.C., Triantafyllou M., Birkhaeuser F.D., et. al.: Combined ultrasmall superparamagnetic particles of iron oxide-enhanced and diffusion-weighted magnetic resonance imaging reliably detect pelvic lymph node metastases in normal-sized nodes of bladder and prostate cancer patients. Eur Urol 2009; 55: pp. 761-769.
77. Derhy S., El Mouhadi S., Ruiz A., et. al.: Non-contrast 3D MR lymphography of retroperitoneal lymphatic aneurysmal dilatation: a continuous spectrum of change from normal variants to cystic lymphangioma. Insights Imaging 2013; 4: pp. 753-758.
78. Arrive L., Derhy S., El Mouhadi S., et. al.: Noncontrast magnetic resonance lymphography. J Reconstr Microsurg 2016; 32: pp. 80-86.
79. Collewet G., Strzelecki M., Mariette F.: Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging 2004; 22: pp. 81-91.
80. Vovk U., Pernuš F., Likar B.: MRI intensity inhomogeneity correction by combining intensity and spatial information. Phys Med Biol 2004; 49: pp. 4119.
81. Vovk U., Pernuš F., Likar B.: A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans Med Imaging 2007; 26: pp. 405-421.
82. Sled J.G., Zijdenbos A.P., Evans A.C.: A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998; 17: pp. 87-97.
83. Tustison N.J., Avants B.B., Cook P.A., et. al.: N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010; 29: pp. 1310-1320.
84. Qiu W., Yuan J., Ukwatta E., et. al.: Dual optimization based prostate zonal segmentation in 3D MR images. Med Image Anal 2014; 18: pp. 660-673.
85. Mahapatra D., Buhmann J.M.: Prostate MRI segmentation using learned semantic knowledge and graph cuts. IEEE Trans Biomed Eng 2014; 61: pp. 756-764.
86. Liao S., Gao Y., Oto A., et. al.: Representation learning: a unified deep learning framework for automatic prostate MR segmentation. Med Image Comput Comput Assist Interv 2013; 16: pp. 254-261.
87. Toth R., Madabhushi A.: Multifeature landmark-free active appearance models: application to prostate MRI segmentation. IEEE Trans Med Imaging 2012; 31: pp. 1638-1650.
88. Litjens G., Debats O., van de Ven W., et. al.: A pattern recognition approach to zonal segmentation of the prostate on MRI. Med Image Comput Comput Assist Interv 2012; 15: pp. 413-420.
89. Qiu W., Yuan J., Ukwatta E., et. al.: Prostate segmentation: an efficient convex optimization approach with axial symmetry using 3-D TRUS and MR images. IEEE Trans Med Imaging 2014; 33: pp. 947-960.
90. Yang M., Li X., Turkbey B., et. al.: Prostate segmentation in MR images using discriminant boundary features. IEEE Trans Biomed Eng 2013; 60: pp. 479-488.
91. Zwiggelaar R., Zhu Y., Williams S.: Semi-automatic segmentation of the prostate.Pattern recognition and image analysis.2003.Springerpp. 1108-1116.
92. Flores-Tapia D., Thomas G., Venugopa N., et. al.: Semi automatic MRI prostate segmentation based on wavelet multiscale products.Engineering in Medicine and Biology Society, 2008 EMBS 2008 30th Annual International Conference of the IEEE.2008.IEEEpp. 3020-3023.
93. Samiee M., Thomas G., Fazel-Rezai R.: Semi-automatic prostate segmentation of MR images based on flow orientation.IEEE International Symposium on Signal Processing and Information Technology, 2006.2006.IEEEpp. 203-207.
94. Cootes T.F., Hill A., Taylor C.J., et. al.: The use of active shape models for locating structures in medical images.Information processing in medical imaging.1993.Springerpp. 33-47.
95. Zhu Y., Williams S., Zwiggelaar R.: A hybrid ASM approach for sparse volumetric data segmentation. Pattern Recognit Image Anal 2007; 17: pp. 252-258.
96. Kass M., Witkin A., Terzopoulos D.: Snakes: active contour models. Int J Comput Vis 1988; 1: pp. 321-331.
97. Chan T.F., Vese L.A.: Active contours without edges. IEEE Trans Image Process 2001; 10: pp. 266-277.
98. Mumford D., Shah J.: Optimal approximations by piecewise smooth functions and associated variational problems. Commun Pure Appl Math 1989; 42: pp. 577-685.
99. Klein S., van der Heide U.A., Lips I.M., et. al.: Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys 2008; 35: pp. 1407-1417.
100. Boykov Y., Veksler O., Zabih R.: Fast approximate energy minimization via graph cuts. IEEE Trans Pattern Anal Mach Intell 2001; 23: pp. 1222-1239.
101. Boykov Y.Y., Jolly M.-P.: Interactive graph cuts for optimal boundary & region segmentation of objects in ND images.2001 Proceedings Eighth IEEE International Conference on Computer Vision, 2001 ICCV.2001.IEEEpp. 105-112.
102. Egger J.: PCG-cut: graph driven segmentation of the prostate central gland. PLoS ONE 2013; 8: pp. e76645.
103. Tian Z., Liu L., Fei B.: A supervoxel-based segmentation for prostate MR images. Proc SPIE 2015; 9413: pp. 941318.
104. Litjens G., Toth R., van de Ven W., et. al.: Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Med Image Anal 2014; 18: pp. 359-373.
105. Ghose S., Oliver A., Marti R., et. al.: A survey of prostate segmentation methodologies in ultrasound, magnetic resonance and computed tomography images. Comput Methods Programs Biomed 2012; 108: pp. 262-287.
106. Oliveira F.P., Tavares J.M.R.: Medical image registration: a review. Comput Methods Biomech Biomed Engin 2014; 17: pp. 73-93.
107. Chappelow J., Madabhushi A., Rosen M., et. al.: Multimodal image registration of ex vivo 4 Tesla MRI with whole mount histology for prostate cancer detection. Proc SPIE 2007; 6512: pp. 65121S.
108. Kalavagunta C., Zhou X., Schmechel S.C., et. al.: Registration of in vivo prostate MRI and pseudo-whole mount histology using Local Affine Transformations guided by Internal Structures (LATIS). J Magn Reson Imaging 2015; 41: pp. 1104-1114.
109. Huisman H.J., Engelbrecht M.R., Barentsz J.O.: Accurate estimation of pharmacokinetic contrast-enhanced dynamic MRI parameters of the prostate. J Magn Reson Imaging 2001; 13: pp. 607-614.
110. Li Q., Sone S., Doi K.: Selective enhancement filters for nodules, vessels, and airway walls in two-and three-dimensional CT scans. Med Phys 2003; 30: pp. 2040-2051.
111. Langer D.L., van der Kwast T.H., Evans A.J., et. al.: Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. J Magn Reson Imaging 2009; 30: pp. 327-334.
112. Murase K.: Efficient method for calculating kinetic parameters using T1-weighted dynamic contrast-enhanced magnetic resonance imaging. Magn Reson Med 2004; 51: pp. 858-862.
113. Lambin P., Rios-Velazquez E., Leijenaar R., et. al.: Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: pp. 441-446.
114. Kumar V., Gu Y., Basu S., et. al.: Radiomics: the process and the challenges. Magn Reson Imaging 2012; 30: pp. 1234-1248.
115. Cameron A., Khalvati F., Haider M., et. al.: MAPS: a quantitative radiomics approach for prostate cancer detection. IEEE Trans Biomed Eng 2015;
116. Wibmer A., Hricak H., Gondo T., et. al.: Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol 2015; 25: pp. 2840-2850.
117. Fehr D., Veeraraghavan H.: Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci USA 2015; 112: pp. E6265-E6273.
118. Khalvati F., Wong A., Haider M.A.: Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med Imaging 2015; 15: pp. 27.
119. Breiman L.: Random forests. Mach Learn 2001; 45: pp. 5-32.
120. Friedman J., Hastie T., Tibshirani R.: Additive logistic regression: a statistical view of boosting (With discussion and a rejoinder by the authors). Ann Stat 2000; 28: pp. 337-407.
121. Baeza-Yates R., Ribeiro-Neto B.: Modern information retrieval.1999.ACM PressNew York
122. Chan I., Wells W., Mulkern R.V., et. al.: Detection of prostate cancer by integration of line-scan diffusion, T2-mapping and T2-weighted magnetic resonance imaging; a multichannel statistical classifier. Med Phys 2003; 30: pp. 2390-2398.
123. Donati O.F., Mazaheri Y., Afaq A., et. al.: Prostate cancer aggressiveness: assessment with whole-lesion histogram analysis of the apparent diffusion coefficient. Radiology 2014; 271: pp. 143-152.
124. Shah V., Pohida T., Turkbey B., et. al.: A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum 2009; 80: pp. 104301.
125. Mazaheri Y., Bokacheva L., Kroon D.J., et. al.: Semi-automatic deformable registration of prostate MR images to pathological slices. J Magn Reson Imaging 2010; 32: pp. 1149-1157.
126. Jacobs M.A., Windham J.P., Soltanian-Zadeh H., et. al.: Registration and warping of magnetic resonance images to histological sections. Med Phys 1999; 26: pp. 1568-1578.
127. Park H., Piert M.R., Khan A., et. al.: Registration methodology for histological sections and in vivo imaging of human prostate. Acad Radiol 2008; 15: pp. 1027-1039.
128. Meyer C.R., Moffat B.A., Kuszpit K.K., et. al.: A methodology for registration of a histological slide and in vivo MRI volume based on optimizing mutual information. Mol Imaging 2006; 5: pp. 16-23.
129. Viswanath S., Bloch B.N., Rosen M., et. al.: Integrating structural and functional imaging for computer assisted detection of prostate cancer on multi-protocol 3 Tesla MRI. Proc Soc Photo Opt Instrum Eng 2009; 7260: pp. 72603I.
130. Gibson E., Gaed M., Gomez J.A., et. al.: 3D prostate histology image reconstruction: quantifying the impact of tissue deformation and histology section location. J Pathol Inform 2013; 4: pp. 31.
131. Gibson E., Gaed M., Gomez J.A., et. al.: 3D prostate histology reconstruction: an evaluation of image-based and fiducial-based algorithms. Med Phys 2013; 40: pp. 093501.
132. Patel P., Chappelow J., Tomaszewski J., et. al.: Spatially weighted mutual information (SWMI) for registration of digitally reconstructed ex vivo whole mount histology and in vivo prostate MRI. Conf Proc IEEE Eng Med Biol Soc 2011; 2011: pp. 6269-6272.
133. McGrath D.M., Vlad R.M., Foltz W.D., et. al.: Technical note: fiducial markers for correlation of whole-specimen histopathology with MR imaging at 7 tesla. Med Phys 2010; 37: pp. 2321-2328.
134. Toth R.J., Shih N., Tomaszewski J.E., et. al.: Histostitcher: an informatics software platform for reconstructing whole-mount prostate histology using the extensible imaging platform framework. J Pathol Inform 2014; 5: pp. 8.
135. Chappelow J., Bloch B.N., Rofsky N., et. al.: Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information. Med Phys 2011; 38: pp. 2005-2018.
136. Orczyk C., Rusinek H., Rosenkrantz A.B., et. al.: Preliminary experience with a novel method of three-dimensional co-registration of prostate cancer digital histology and in vivo multiparametric MRI. Clin Radiol 2013; 68: pp. e652-e658.
137. Lv D., Guo X., Wang X., et. al.: Computerized characterization of prostate cancer by fractal analysis in MR images. J Magn Reson Imaging 2009; 30: pp. 161-168.
138. Rouviere O., Papillard M., Girouin N., et. al.: Is it possible to model the risk of malignancy of focal abnormalities found at prostate multiparametric MRI?. Eur Radiol 2012; 22: pp. 1149-1157.
139. Meyer C., Ma B., Kunju L.P., et. al.: Challenges in accurate registration of 3-D medical imaging and histopathology in primary prostate cancer. Eur J Nucl Med Mol Imaging 2013; 40: pp. 72-78.
140. Viswanath S., Bloch B.N., Genega E., et. al.: A comprehensive segmentation, registration, and cancer detection scheme on 3 Tesla in vivo prostate DCE-MRI. Med Image Comput Comput Assist Interv 2008; 11: pp. 662-669.
141. Liu X., Langer D.L., Haider M.A., et. al.: Prostate cancer segmentation with simultaneous estimation of Markov random field parameters and class. IEEE Trans Med Imaging 2009; 28: pp. 906-915.
142. Artan Y., Haider M.A., Langer D.L., et. al.: Prostate cancer localization with multispectral MRI using cost-sensitive support vector machines and conditional random fields. IEEE Trans Image Process 2010; 19: pp. 2444-2455.
143. Vos P.C., Hambrock T., Barenstz J.O., et. al.: Computer-assisted analysis of peripheral zone prostate lesions using T2-weighted and dynamic contrast enhanced T1-weighted MRI. Phys Med Biol 2010; 55: pp. 1719-1734.
144. Viswanath S., Bloch B.N., Chappelow J., et. al.: Enhanced multi-protocol analysis via intelligent supervised embedding (EMPrAvISE): detecting prostate cancer on multi-parametric MRI. Proc SPIE Int Soc Opt Eng 2011; 7963: pp. 79630u.
145. Lopes R., Ayache A., Makni N., et. al.: Prostate cancer characterization on MR images using fractal features. Med Phys 2011; 38: pp. 83-95.
146. Sung Y.S., Kwon H.J., Park B.W., et. al.: Prostate cancer detection on dynamic contrast-enhanced MRI: computer-aided diagnosis versus single perfusion parameter maps. AJR Am J Roentgenol 2011; 197: pp. 1122-1129.
147. Artan Y., Yetik I.S.: Prostate cancer localization using multiparametric MRI based on semi-supervised techniques with automated seed initialization. IEEE Trans Inf Technol Biomed 2012; 16: pp. 1313-1323.
148. Tiwari P., Kurhanewicz J., Madabhushi A.: Multi-kernel graph embedding for detection, Gleason grading of prostate cancer via MRI/MRS. Med Image Anal 2013; 17: pp. 219-235.
149. Ginsburg S.B., Viswanath S.E., Bloch B.N., et. al.: Novel PCA-VIP scheme for ranking MRI protocols and identifying computer-extracted MRI measurements associated with central gland and peripheral zone prostate tumors. J Magn Reson Imaging 2015; 41: pp. 1383-1393.
150. Niaf E., Flamary R., Rouviere O., et. al.: Kernel-based learning from both qualitative and quantitative labels: application to prostate cancer diagnosis based on multiparametric MR imaging. IEEE Trans Image Process 2014; 23: pp. 979-991.
151. Zhao K., Wang C., Hu J., et. al.: Prostate cancer identification: quantitative analysis of T2-weighted MR images based on a back propagation artificial neural network model. Sci China Life Sci 2015; 58: pp. 666-673.
152. Saman D.M., Lemieux A.M., Nawal Lutfiyya M., et. al.: A review of the current epidemiology and treatment options for prostate cancer. Dis Mon 2014; 60: pp. 150-154.
153. Keyes M., Crook J., Morton G., et. al.: Treatment options for localized prostate cancer. Can Fam Physician 2013; 59: pp. 1269-1274.
154. Oto A., Yang C., Kayhan A., et. al.: Diffusion-weighted and dynamic contrast-enhanced MRI of prostate cancer: correlation of quantitative MR parameters with Gleason score and tumor angiogenesis. AJR Am J Roentgenol 2011; 197: pp. 1382-1390.
155. Anwar S.S., Anwar Khan Z., Shoaib Hamid R., et. al.: Assessment of apparent diffusion coefficient values as predictor of aggressiveness in peripheral zone prostate cancer: comparison with Gleason score. ISRN Radiol 2014; 2014: pp. 263417.
156. Hambrock T., Somford D.M., Huisman H.J., et. al.: Relationship between apparent diffusion coefficients at 3.0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology 2011; 259: pp. 453-461.
157. Yamamura J., Salomon G., Buchert R., et. al.: MR imaging of prostate cancer: diffusion weighted imaging and (3D) hydrogen 1 (H) MR spectroscopy in comparison with histology. Radiol Res Pract 2011; 2011: pp. 616852.
158. Gibbs P., Liney G.P., Pickles M.D., et. al.: Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 Tesla. Invest Radiol 2009; 44: pp. 572-576.
159. Anderson A.W., Xie J., Pizzonia J., et. al.: Effects of cell volume fraction changes on apparent diffusion in human cells. Magn Reson Imaging 2000; 18: pp. 689-695.
160. Tanimoto K., Yoshikawa K., Obata T., et. al.: Role of glucose metabolism and cellularity for tumor malignancy evaluation using FDG-PET/CT and MRI. Nucl Med Commun 2010; 31: pp. 604-609.
161. Kobus T., Hambrock T., Hulsbergen-van de Kaa C.A., et. al.: In vivo assessment of prostate cancer aggressiveness using magnetic resonance spectroscopic imaging at 3 T with an endorectal coil. Eur Urol 2011; 60: pp. 1074-1080.
162. Zakian K.L., Sircar K., Hricak H., et. al.: Correlation of proton MR spectroscopic imaging with Gleason score based on step-section pathologic analysis after radical prostatectomy. Radiology 2005; 234: pp. 804-814.
163. van Asten J.J., Cuijpers V., Hulsbergen-van de Kaa C., et. al.: High resolution magic angle spinning NMR spectroscopy for metabolic assessment of cancer presence and Gleason score in human prostate needle biopsies. MAGMA 2008; 21: pp. 435-442.
164. Garcia-Martin M.L., Adrados M., Ortega M.P., et. al.: Quantitative (1) H MR spectroscopic imaging of the prostate gland using LCModel and a dedicated basis-set: correlation with histologic findings. Magn Reson Med 2011; 65: pp. 329-339.
165. Scheenen T.W., Heijmink S.W., Roell S.A., et. al.: Three-dimensional proton MR spectroscopy of human prostate at 3 T without endorectal coil: feasibility. Radiology 2007; 245: pp. 507-516.
166. Shukla-Dave A., Hricak H., Ishill N.M., et. al.: Correlation of MR imaging and MR spectroscopic imaging findings with Ki-67, phospho-Akt, and androgen receptor expression in prostate cancer. Radiology 2009; 250: pp. 803-812.
167. Lee D.H., Koo K.C., Lee S.H., et. al.: Tumor lesion diameter on diffusion weighted magnetic resonance imaging could help predict insignificant prostate cancer in patients eligible for active surveillance: preliminary analysis. J Urol 2013; 190: pp. 1213-1217.
168. Epstein J.I., Allsbrook W.C., Amin M.B., et. al.: The 2005 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol 2005; 29: pp. 1228-1242.
169. Cam K., Yucel S., Turkeri L., et. al.: Accuracy of transrectal ultrasound guided prostate biopsy: histopathological correlation to matched prostatectomy specimens. Int J Urol 2002; 9: pp. 257-260.
170. Quann P., Jarrard D.F., Huang W.: Current prostate biopsy protocols cannot reliably identify patients for focal therapy: correlation of low-risk prostate cancer on biopsy with radical prostatectomy findings. Int J Clin Exp Pathol 2010; 3: pp. 401-407.
171. Djavan B., Ravery V., Zlotta A., et. al.: Prospective evaluation of prostate cancer detected on biopsies 1, 2, 3 and 4: when should we stop?. J Urol 2001; 166: pp. 1679-1683.
172. Futterer J.J., Barentsz J.O.: MRI-guided and robotic-assisted prostate biopsy. Curr Opin Urol 2012; 22: pp. 316-319.
173. Moore C.M., Robertson N.L., Arsanious N., et. al.: Image-guided prostate biopsy using magnetic resonance imaging-derived targets: a systematic review. Eur Urol 2013; 63: pp. 125-140.
174. Xu H., Lasso A., Guion P., et. al.: Accuracy analysis in MRI-guided robotic prostate biopsy. Int J Comput Assist Radiol Surg 2013; 8: pp. 937-944.
175. Zamecnik P., Schouten M.G., Krafft A.J., et. al.: Automated real-time needle-guide tracking for fast 3-T MR-guided transrectal prostate biopsy: a feasibility study. Radiology 2014; 273: pp. 879-886.
176. Fiard G., Hohn N., Descotes J.L., et. al.: Targeted MRI-guided prostate biopsies for the detection of prostate cancer: initial clinical experience with real-time 3-dimensional transrectal ultrasound guidance and magnetic resonance/transrectal ultrasound image fusion. Urology 2013; 81: pp. 1372-1378.
177. Xu S., Kruecker J., Turkbey B., et. al.: Real-time MRI-TRUS fusion for guidance of targeted prostate biopsies. Comput Aided Surg 2008; 13: pp. 255-264.
178. Mitra J., Ghose S., Sidibe D., et. al.: Joint probability of shape and image similarities to retrieve 2D TRUS-MR slice correspondence for prostate biopsy. Conf Proc IEEE Eng Med Biol Soc 2012; 2012: pp. 5416-5419.
179. Wang S., Burtt K., Turkbey B., et. al.: Computer aided-diagnosis of prostate cancer on multiparametric MRI: a technical review of current research. Biomed Res Int 2014; 2014: pp. 789561.
180. Fei B.W., Lee Z.H., Boll D.T., et. al.: Image registration and fusion for interventional MRI guided thermal ablation of the prostate cancer.Ellis R.E.Peters T.M.Medical image computing and computer-assisted intervention—MICCAI 2003, Pt 2.2003.pp. 364-372.
181. Akbari H., Fei B.W.: 3D ultrasound image segmentation using wavelet support vector machines. Med Phys 2012; 39: pp. 2972-2984.
182. Fei B.W., Lee Z.H., Boll D.T., et. al.: Registration and fusion of SPECT, high-resolution MRI, and interventional MRI for thermal ablation of prostate cancer. IEEE Trans Nucl Sci 2004; 51: pp. 177-183.
183. Fei B.W., Lee Z.H., Duerk J.L., et. al.: Image registration for interventional MRI guided procedures: interpolation methods, similarity measurements, and applications to the prostate.Gee J.C.Maintz J.B.A.Vannier M.W.Biomedical image registration.2003.pp. 321-329.
184. Fei B.W., Ng W.S., Chauhan S., et. al.: The safety issues of medical robotics. Reliab Eng Syst Saf 2001; 73: pp. 183-192.
185. Fei B.W., Wang H., Meyers J.D., et. al.: High-field magnetic resonance imaging of the response of human prostate cancer to Pc 4-based photodynamic therapy in an animal model. Lasers Surg Med 2007; 39: pp. 723-730.
186. Fei B.W., Wang H.S., Wu C.Y., et. al.: Choline PET for monitoring early tumor response to photodynamic therapy. J Nucl Med 2010; 51: pp. 130-138.
187. Fei B.W., Yang X.F., Nye J.A., et. al.: MR/PET quantification tools: registration, segmentation, classification, and MR-based attenuation correction. Med Phys 2012; 39: pp. 6443-6454.
188. Wang H.S., Fei B.W.: Diffusion-weighted MRI for monitoring tumor response to photodynamic therapy. J Magn Reson Imaging 2010; 32: pp. 409-417.
189. Wang H.S., Fei B.W.: An MR image-guided, voxel-based partial volume correction method for PET images. Med Phys 2012; 39: pp. 179-194.
190. Wang H.S., Fei B.W.: Nonrigid point registration for 2D curves and 3D surfaces and its various applications. Phys Med Biol 2013; 58: pp. 4315-4330.
191. Song I., Kim C.K., Park B.K., et. al.: Assessment of response to radiotherapy for prostate cancer: value of diffusion-weighted MRI at 3 T. AJR Am J Roentgenol 2010; 194: pp. W477-W482.
192. Padhani A.R., Makris A., Gall P., et. al.: Therapy monitoring of skeletal metastases with whole-body diffusion MRI. J Magn Reson Imaging 2014; 39: pp. 1049-1078.
193. Graham T.J., Box G., Tunariu N., et. al.: Preclinical evaluation of imaging biomarkers for prostate cancer bone metastasis and response to cabozantinib. J Natl Cancer Inst 2014; 106: pp. dju033.
194. Lee K.C., Bradley D.A., Hussain M., et. al.: A feasibility study evaluating the functional diffusion map as a predictive imaging biomarker for detection of treatment response in a patient with metastatic prostate cancer to the bone. Neoplasia 2007; 9: pp. 1003-1011.
195. Sosna J., Pedrosa I., Dewolf W.C., et. al.: MR imaging of the prostate at 3 Tesla: comparison of an external phased-array coil to imaging with an endorectal coil at 1.5 Tesla. Acad Radiol 2004; 11: pp. 857-862.
196. Turkbey B., Merino M.J., Gallardo E.C., et. al.: Comparison of endorectal coil and nonendorectal coil T2W and diffusion-weighted MRI at 3 Tesla for localizing prostate cancer: correlation with whole-mount histopathology. J Magn Reson Imaging 2014; 39: pp. 1443-1448.
197. Litjens G.J., Barentsz J.O., Karssemeijer N., et. al.: Clinical evaluation of a computer-aided diagnosis system for determining cancer aggressiveness in prostate MRI. Eur Radiol 2015; 25: pp. 3187-3199.
198. Matulewicz L., Jansen J.F., Bokacheva L., et. al.: Anatomic segmentation improves prostate cancer detection with artificial neural networks analysis of 1H magnetic resonance spectroscopic imaging. J Magn Reson Imaging 2014; 40: pp. 1414-1421.
199. Huang P.W., Lee C.H.: Automatic classification for pathological prostate images based on fractal analysis. IEEE Trans Med Imaging 2009; 28: pp. 1037-1050.
200. Sonn G.A., Margolis D.J., Marks L.S.: Target detection: magnetic resonance imaging-ultrasound fusion–guided prostate biopsy. Urol Oncol 2014; 32: pp. 903-911.
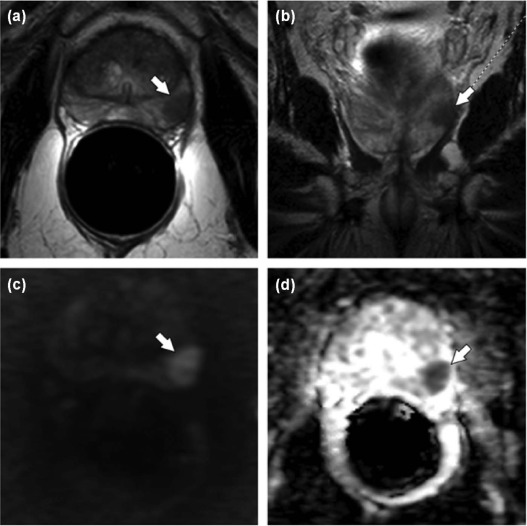
![Figure 3, Dynamic contrast-enhanced magnetic resonance imaging (MRI) (dynamic contrast-enhanced imaging [DCE-MRI]) of the prostate. (a) Axial T1 gradient echo (GRE) sequence unenhanced image. After contrast agent administration, an area with early enhancement is seen on the right in the peripheral zone ( b ), region of interest [ROI1]) with significant washout in the late-phase image (c) . The curve ( red ) with early enhancement is a typical finding in the case of prostate cancer, whereas healthy prostate tissue is characterized by a steady slow enhancement ( green ). High transport constants K trans (e) and k ep (f) can confirm suspicion of prostate cancer. Prostate adenocarcinoma with a Gleason score of 4 + 5 = 9 was diagnosed after prostatectomy](https://storage.googleapis.com/dl.dentistrykey.com/clinical/ComputeraidedDetectionofProstateCancerwithMRI/2_1s20S1076633216300095.jpg)

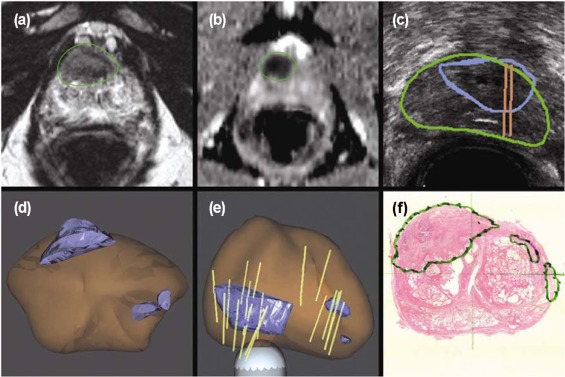
![Figure 11, Registration between magnetic resonance imaging (MRI) and histology. Top : Workflow for pathology-multiparametric (mp)-MRI registration in a surgical three-dimensional (3D) space. Bottom : 3D deformable registration of virtual whole-mount histology (1), fresh specimen (2), T2-weighted MRI (3), perfusion (4), and diffusion (5) sequences (apparent diffusion coefficient [ADC]) applied to prostate cancer](https://storage.googleapis.com/dl.dentistrykey.com/clinical/ComputeraidedDetectionofProstateCancerwithMRI/10_1s20S1076633216300095.jpg)