Rationale and Objectives
The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI) is the largest publicly available computed tomography (CT) image reference data set of lung nodules. In this article, a comprehensive data analysis of the data set and a uniform data model are presented with the purpose of facilitating potential researchers to have an in-depth understanding to and efficient use of the data set in their lung cancer–related investigations.
Materials and Methods
A uniform data model was designed for representation and organization of various types of information contained in different source data files. A software tool was developed for the processing and analysis of the database, which 1) automatically aligns and graphically displays the nodule outlines marked manually by radiologists onto the corresponding CT images; 2) extracts diagnostic nodule characteristics annotated by radiologists; 3) calculates a variety of nodule image features based on the outlines of nodules, including diameter, volume, and degree of roundness, and so forth; 4) integrates all the extracted nodule information into the uniform data model and stores it in a common and easy-to-access data format; and 5) analyzes and summarizes various feature distributions of nodules in several different categories. Using this data processing and analysis tool, all 1018 CT scans from the data set were processed and analyzed for their statistical distribution.
Results
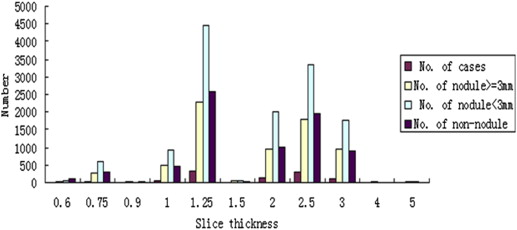
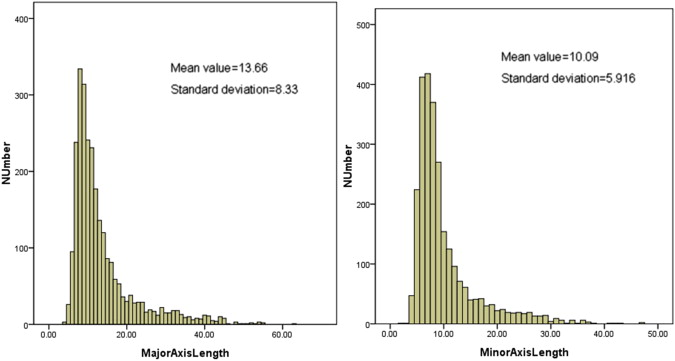
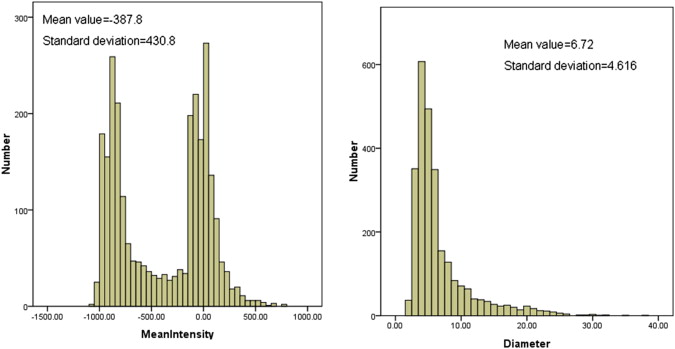
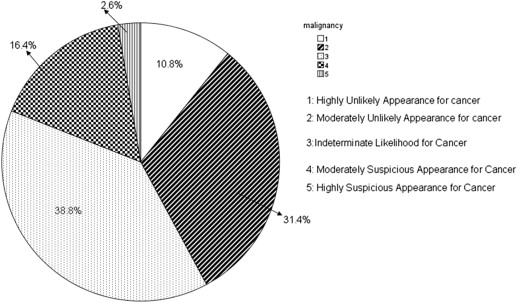
The information contained in different source data files with different formats was extracted and integrated into a new and uniform data model. Based on the new data model, the statistical distributions of nodules in terms of nodule geometric features and diagnostic characteristics were summarized. In the LIDC/IDRI data set, 2655 nodules ≥3 mm, 5875 nodules <3 mm, and 7411 non-nodules are identified, respectively. Among the 2655 nodules, 1) 775, 488, 481, and 911 were marked by one, two, three, or four radiologists, respectively; 2) most of nodules ≥3 mm (85.7%) have a diameter <10.0 mm with the mean value of 6.72 mm; and 3) 10.87%, 31.4%, 38.8%, 16.4%, and 2.6% of nodules were assessed with a malignancy score of 1, 2, 3, 4, and 5, respectively.
Conclusions
This study demonstrates the usefulness of the proposed software tool to the potential users for an in-depth understanding of the LIDC/IDRI data set, therefore likely to be beneficial to their future investigations. The analysis results also demonstrate the distribution diversity of nodules characteristics, therefore being useful as a reference resource for assessing the performance of a new and existing nodule detection and/or segmentation schemes.
Collecting a large number of computed tomography (CT) scans and developing rigorous ground truth is essential for developing computer-aided detection (CAD) or diagnosis methods for lung nodules. In 2000, the National Cancer Institute (NCI) initiated a multi-institutional effort to establish a publicly available reference database named the Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI) . The motivation was to allow researchers around the world to share a large and diverse image data set for various purposes, especially for developing, training, and evaluating CAD approaches developed by different research groups. By 2011, the LIDC/IDRI database contained 1018 CT scans in total. Now, it has been widely used in a number of researches related to lung cancer, such as lung nodule detection or classification, either as training and/or testing data set in developing and evaluating computer-aided detection (CAD) or computer-aided diagnosis (CADx) systems .
Efficient and effective use of the LIDC/IDRI data set is, however, still affected by several barriers. One of the major barriers is the absence of in-depth analysis of the lung nodules data. In the LIDC/IDRI data set, each case includes images from a clinical thoracic CT scan and an associated Extensive Markup Language (XML) file. Each XML file records the outlines of nodules and their diagnosis assessments given by four experienced thoracic radiologists. The distribution characteristics of the nodules in the annotation files have not been fully analyzed and made available to researchers. This resulted in several potential difficulties to the researchers who use the data set because the performance and reliability of a CAD scheme heavily depend on the characteristics of the training and testing data set. Solid understanding of the diversity and characteristics of the nodules used in investigates is important for researchers to develop and evaluate CAD approaches. For example, the percentage of nodules in terms of size, shape characteristics, and CT slice thickness are very important factors for developing an efficient CAD scheme for lung nodule detection. A global analysis of the diagnosis assessment ranks provided by the radiology experts is also critical for developing CAD approaches in discriminating malignancy nodules from benign nodules. Therefore, developing an efficient and reliable method for automatic analysis and presentation of the characteristics of the LIDC/IDRI will be beneficial to many researchers.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and methods
Data
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Methods
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
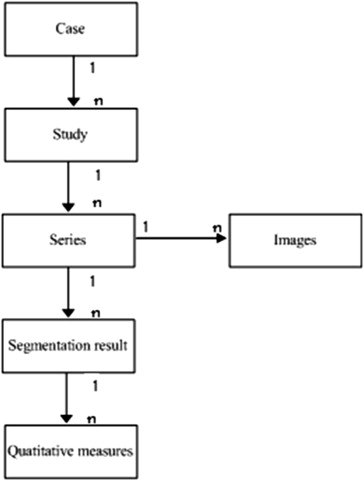
Design of Data Model
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Extraction of Information from the LIDC/IDRI Database
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Derivation of Quantitative Features
Get Radiology Tree app to read full this article<
Table 1
All Calculated Features of Nodules
Shape Features Size Features Intensity Features Circularity
Roughness
Elongation
Compactness
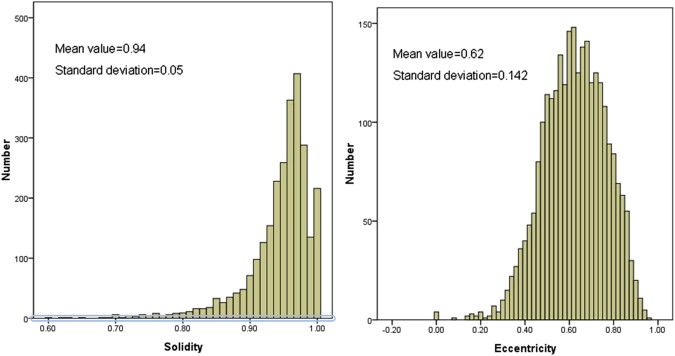
Eccentricity
Solidity
Extent
Sphericity Area
Convex area
Filled area
Perimeter
Convex perimeter
Equiv diameter
Major axis length
Minor axis length
Volume
Equivalent diameter Minimum intensity
Maximum intensity
Mean intensity
Intensity standard
Intensity difference
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Equivalent diameter=23V4π−−−√3 Equivalent diameter
=
2
3
V
4
π
3
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Circularity=4π⋅AP2 C
i
r
c
u
l
a
r
i
t
y
=
4
π
·
A
P
2
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Quantitative Analysis
Get Radiology Tree app to read full this article<
Results
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 2
The Distribution of the Number of Nodules and Non-nodules Marked by Different Number of Radiologists
No. of Radiologists No. of Nodules (Nodules ≥3 mm) No. of Nodules (Nodules <3 mm) No. of Non-nodules 1 775 (29.2) 1468 (25.0) 2232 (30.1) 2 488 (18.4) 1177 (20.0) 2260 (30.5) 3 481 (18.1) 1106 (18.8) 1705 (23.0) 4 911 (34.3) 2124 (36.2) 1214 (16.4)
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
Acknowledgments
Get Radiology Tree app to read full this article<
References
1. Armato S.G., et. al.: The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011; 38: pp. 915-932.
2. Clarke L.P., et. al.: National Cancer Institute initiative: lung image database resource for imaging research. Acad Radiol 2001; 8: pp. 447-450.
3. McNitt-Gray M.F., et. al.: The Lung Image Database Consortium (LIDC) data collection process for nodule detection and annotation. Acad Radiol 2007; 14: pp. 1464-1474.
4. Schilham A.M.R., Van Ginneken B., Loog M.: A computer-aided diagnosis system for detection of lung nodules in chest radiographs with an evaluation on a public database. Med Image Anal 2006; 10: pp. 247-258.
5. Retico A., et. al.: Lung nodule detection in low-dose and thin-slice computed tomography. Comput Biol Med 2008; 38: pp. 525-534.
6. Hardie R.C., et. al.: Performance analysis of a new computer aided detection system for identifying lung nodules on chest radiographs. Med Image Anal 2008; 12: pp. 240-258.
7. Messay T., Hardie R.C., Rogers S.K.: A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med Image Anal 2010; 14: pp. 390-406.
8. Reeves A.P., Kostis W.J.: Computer-aided diagnosis for lung cancer. Radiol Clin North Am 2000; 38: pp. 497-509.
9. Iwano S., et. al.: Computer-aided diagnosis: a shape classification of pulmonary nodules imaged by high-resolution CT. Comput Med Imaging Graph 2005; 29: pp. 565-570.
10. Way T.W., Hadjiiski L.M., Sahiner B., et. al.: Computer-aided diagnosis of pulmonary nodules on CT scans: segmentation and classification using 3D active contours. Med Phys 2006; 33: pp. 2323.
11. Lin H., Chen Z., Wang W.: A pulmonary nodule view system for the Lung Image Database Consortium (LIDC). Acad Radiol 2011; 18: pp. 1181-1185.
12. Reeves A.P., Biancardi A.M., Apanasovich T.V., et. al.: The Lung Image Database Consortium (LIDC): a comparison of different size metrics for pulmonary nodule measurements. Acad Radiol 2007; 14: pp. 1475-1485.
13. Ross J.C., Miller J.V., Turner W.D., et. al.: An analysis of early studies released by the Lung Imaging Database Consortium (LIDC). Acad Radiol 2007; 14: pp. 1382-1388.
14. Tan J., Pu J., Zheng B., et. al.: Computerized comprehensive data analysis of Lung Imaging Database Consortium (LIDC). Med Phys 2010; 37: pp. 3802-3808.
15. Available at: https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI ; Accessed October 20, 2013.