Rationale and Objectives
The Liver Imaging Reporting and Data System (LI-RADS) can enhance communication between radiologists and clinicians if applied consistently. We identified an institutional need to improve liver imaging report standardization and developed handheld and desktop software to serve this purpose.
Materials and Methods
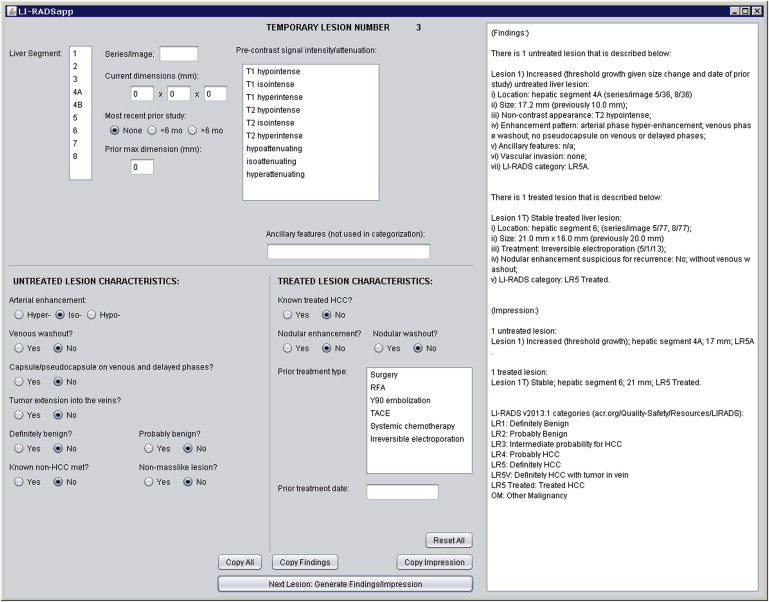
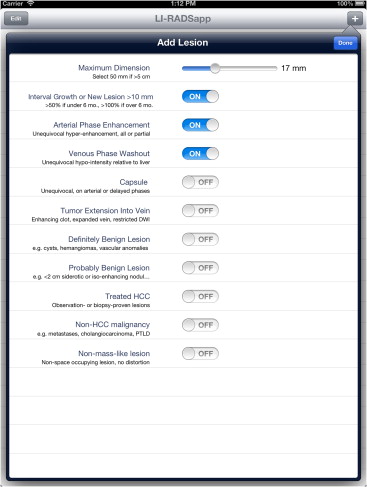
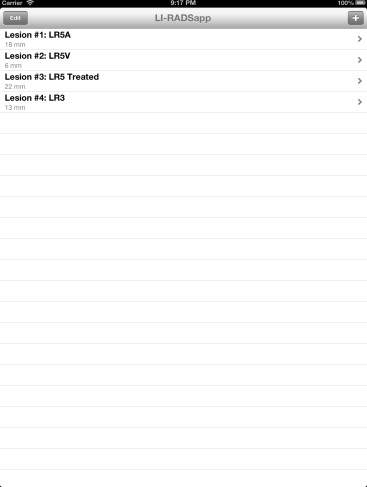
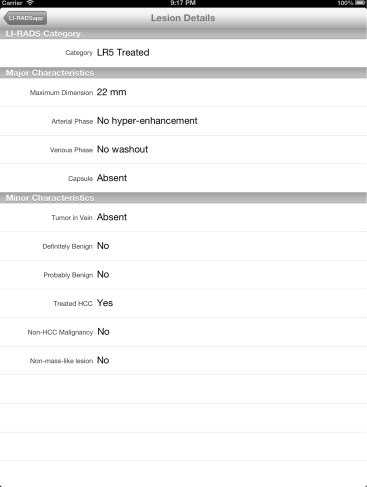
We developed two complementary applications that implement the LI-RADS schema. A mobile application for iOS devices written in the Objective-C language allows for rapid characterization of hepatic observations under a variety of circumstances. A desktop application written in the Java language allows for comprehensive observation characterization and standardized report text generation. We chose the applications’ languages and feature sets based on the computing resources of target platforms, anticipated usage scenarios, and ease of application installation, deployment, and updating.
Results
Our primary results are the publication of the core source code implementing the LI-RADS algorithm and the availability of the applications for use worldwide via our website, http://www.liradsapp.com/ . The Java application is free open-source software that can be integrated into nearly any vendor’s reporting system. The iOS application is distributed through Apple’s iTunes App Store. Observation categorizations of both programs have been manually validated to be correct. The iOS application has been used to characterize liver tumors during multidisciplinary conferences of our institution, and several faculty members, fellows, and residents have adopted the generated text of Java application into their diagnostic reports.
Conclusions
Although these two applications were developed for the specific reporting requirements of our liver tumor service, we intend to apply this development model to other diseases as well. Through semiautomated structured report generation and observation characterization, we aim to improve patient care while increasing radiologist efficiency.
The radiology report is an essential part of the medical record, encompassing several roles. Although its primary function is to communicate important findings and to guide clinical decision-making, the radiology report is also an informational resource for medicolegal, research-related, and financial inquiries . Nonetheless, the conventional approach to radiology reporting has included few constraints on lexical or editorial choices. Consequently, radiology reports are highly inconsistent in terms of their accuracy, completeness, and clarity .
The adoption of uniform subspecialty lexicons and report formats may improve the usability of radiology reports. Perhaps, the most successful effort has been the Breast Imaging Reporting and Data System (BI-RADS), which was developed by the American College of Radiology (ACR) to reduce variability in the assessment, reporting, and management of findings in breast tissue . Its initial implementation led to substantial improvements in the clarity and consistency of mammography reports , thereby prompting the Food and Drug Administration (FDA) to mandate its adoption by breast imagers nationwide. Furthermore, by facilitating the usage of clinical data embedded in mammography reports, the BI-RADS schema has enhanced clinical research and quality assurance initiatives within the field of breast imaging .
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Figure 1
LI-RADS v2013.1 observation categorization flowchart. HCC, hepatocellular carcinoma; LI-RADS, Liver Imaging Reporting and Data System. (LR1, LR2, LR3, LR4, and LR5 represent LI-RADS categories).
Reprinted with permission of the American College of Radiology. No other representation of this material is authorized without expressed, written permission from the American College of Radiology.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and methods
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Results
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Appendix 1
Java lesion characterization function
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Appendix 2
Objective-C lesion characterization method
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Appendix 3
Proposed Java function implementing API for use with CAD software
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Appendix 4
Sample generated report text from Java application
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Duszak R., Nossal M., Schofield L., et. al.: Physician documentation deficiencies in abdominal ultrasound reports: frequency, characteristics, and financial impact. J Am Coll Radiol 2012; 9: pp. 403-408.
2. Thorwarth W.T.: Get paid for what you do: dictation patterns and impact on billing accuracy. J Am Coll Radiol 2005; 2: pp. 665-669.
3. Lucey L.L., Kushner D.C., American College of Radiology: The ACR guideline on communication: to be or not to be, that is the question. J Am Coll Radiol 2010; 7: pp. 109-114.
4. Duszak R.: Dictating for dollars (and compliance, too). J Am Coll Radiol 2004; 1: pp. 874-875.
5. Johnson A.J., Chen M.Y., Zapadka M.E., et. al.: Radiology report clarity: a cohort study of structured reporting compared with conventional dictation. J Am Coll Radiol 2010; 7: pp. 501-506.
6. Robinson P.J.: Radiology’s Achilles’ heel: error and variation in the interpretation of the Rontgen image. Br J Radiol 1997; 70: pp. 1085-1098.
7. Khorasani R., Bates D.W., Teeger S., et. al.: Is terminology used effectively to convey diagnostic certainty in radiology reports?. Acad Radiol 2003; 10: pp. 685-688.
8. Kong A., Barnett G.O., Mosteller F., et. al.: How medical professionals evaluate expressions of probability. N Engl J Med 1986; 315: pp. 740-744.
9. American College of Radiology: . Breast Imaging–Reporting and Data System (BI-RADS).4th ed.2003.American College of RadiologyReston, VA
10. Lazarus E., Mainiero M.B., Schepps B., et. al.: BI-RADS lexicon for US and mammography: interobserver variability and positive predictive value. Radiology 2006; 239: pp. 385-391.
11. Taplin S.H., Ichikawa L.E., Kerlikowske K., et. al.: Concordance of breast imaging reporting and data system assessments and management recommendations in screening mammography. Radiology 2002; 222: pp. 529-535.
12. Geller B.M., Barlow W.E., Ballard-Barbash R., et. al.: Use of the American College of Radiology BI-RADS to report on the mammographic evaluation of women with signs and symptoms of breast disease. Radiology 2002; 222: pp. 536-542.
13. Burnside E.S., Sickles E.A., Bassett L.W., et. al.: The ACR BI-RADS experience: learning from history. J Am Coll Radiol 2009; 6: pp. 851-860.
14. Purysko A.S., Remer E.M., Coppa C.P., et. al.: LI-RADS: a case-based review of the new categorization of liver findings in patients with end-stage liver disease. Radiographics 2012; 32: pp. 1977-1995.
15. American College of Radiology. Quality and safety resources: Liver Imaging–Reporting and Data System. Available at: http://www.acr.org/Quality-Safety/Resources/LIRADS . Accessed August 22, 2013.;
16. Tang A., Cruite I., Sirlin C.B.: Toward a standardized system for hepatocellular carcinoma diagnosis using computed tomography and MRI. Exp Rev Gastroenterol Hepatol 2013; 7: pp. 269-279.
17. Freeman R.B., Mithoefer A., Ruthazer R., et. al.: Optimizing staging for hepatocellular carcinoma before liver transplantation: a retrospective analysis of the UNOS/OPTN database. Liver Transpl 2006; 12: pp. 1504-1511.
18. Hertl M., Cosimi A.B.: Liver transplantation for malignancy. Oncologist 2005; 10: pp. 269-281.