Rationale and Objectives
The aim of this study was to develop a personalized training system using the Lung Image Database Consortium (LIDC) and Image Database resource Initiative (IDRI) Database, because collecting, annotating, and marking a large number of appropriate computed tomography (CT) scans, and providing the capability of dynamically selecting suitable training cases based on the performance levels of trainees and the characteristics of cases are critical for developing a efficient training system.
Materials and Methods
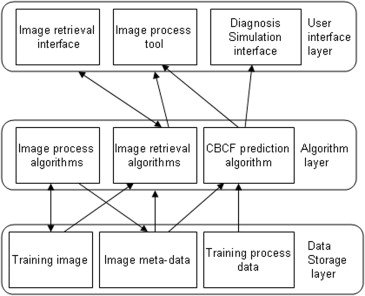
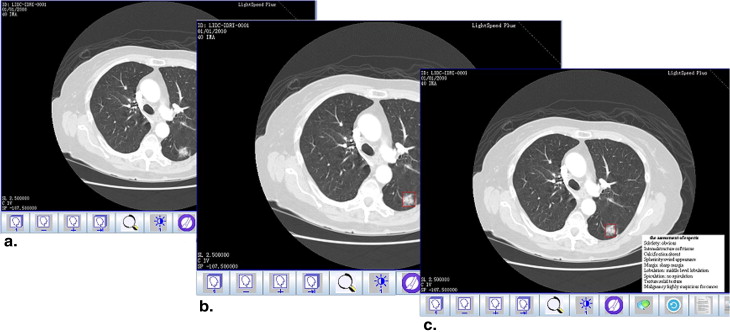
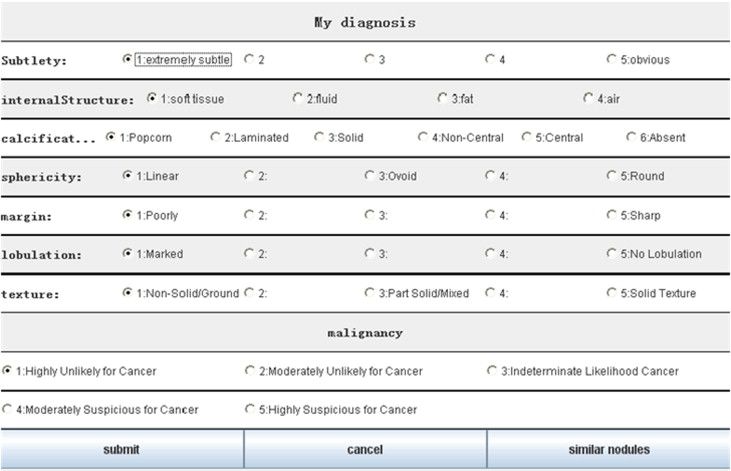
A novel approach is proposed to develop a personalized radiology training system for the interpretation of lung nodules in CT scans using the Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI) database, which provides a Content-Boosted Collaborative Filtering (CBCF) algorithm for predicting the difficulty level of each case of each trainee when selecting suitable cases to meet individual needs, and a diagnostic simulation tool to enable trainees to analyze and diagnose lung nodules with the help of an image processing tool and a nodule retrieval tool.
Results
Preliminary evaluation of the system shows that developing a personalized training system for interpretation of lung nodules is needed and useful to enhance the professional skills of trainees.
Conclusions
The approach of developing personalized training systems using the LIDC/IDRL database is a feasible solution to the challenges of constructing specific training program in terms of cost and training efficiency.
Lung cancer has been the most mortal cancer for both men and women in the last two decades . For example, in 2013, lung cancer is expected to account for 26% of all female cancer deaths and 28% of all male cancer deaths in America according to the 2013 Cancer Statistics established by the American Cancer Society . A number of studies suggest that early detection and diagnosis is the most promising means of increasing the survival rate of patients . Therefore, a significant degree of research has been undertaken to improve the diagnosis and detection accuracy by radiologists. A variety of computer-aided detection (CAD) systems were developed to assist radiologists in detecting lung nodules in computed tomography (CT) scans , and a number of computer-aided diagnosis (CADx) approaches were proposed to assist radiologists in distinguishing malignant nodules from benign ones . The CAD systems being developed for lung cancer detection and classification may require training and evaluation based on CT images. It is also useful to assess the performance of different CAD systems developed by different research groups and to verify their potential clinical utility. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI) have established a reference database, called the LIDC/IDRI database, publicly available to the medical imaging research community. This initiative was sponsored by the National Cancer Institute, further advanced by the Foundation for the National Institutes of Health, and accompanied by the Food and Drug Administration through active participation .
In addition to the assessment of the performance of the various CAD methods, the LIDC/IDRI database has also inspired other research with a variety of applications . For example, Michael et al. created the Content-Based Image Retrieval (CBIR) framework to retrieve images of similar nodules among CT images of pulmonary nodules from the collection provided by the LIDC. They compared three feature extraction methods: 1) Haralick co-occurrence, 2) Gabor filters, and 3) Markov random field. The results showed that the Gabor and Markov descriptors perform better than does the Haralick co-occurrence method at retrieving similar nodules. To bridge the semantic gap between radiologists’ ratings and image features, Dasovich et al. researched the relationship between semantic and content-based similarity using LIDC. They developed a conceptual-based similarity model derived from content-based similarity to improve CBIR. The potential value of these resources is still under active exploration.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Methods
Get Radiology Tree app to read full this article<
Data
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Prediction Algorithm for Personalized Training
Get Radiology Tree app to read full this article<
The Definition of Difficulty Level of a Case for a Given Trainee
Get Radiology Tree app to read full this article<
Ru,i=|lu,i−li||R|, R
u
,
i
=
|
l
u
,
i
−
l
i
|
|
R
|
,
where l__u,i is the assessment of the likelihood of malignancy of case i by trainee u ; l__i is the “gold standard” of case i ; and |R| denotes the rating scale. In this article, |R| is equal to 1. For example, if a trainee’s assessment of a case is 60% of the probability of a lesion being malignant and the “gold standard” is 75%, then the difficulty level of this case for this trainee is 15%. And if another trainee’s assessment of the same case is 70%, then the difficulty level of this case for him or her is 5%.
Get Radiology Tree app to read full this article<
Content-Boosted Collaborative Filtering
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Step 1: generating pseudoratings matrix using a content-based predictor
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Sim(i,j)=cos(Fi,Fj)=Fi⋅Fj∥Fi∥∥Fj∥=∑mk=1Fik×Fjk∑mk=1(Fim)2√×∑mk=1(Fjm)2√, S
i
m
(
i
,
j
)
=
cos
(
F
i
,
F
j
)
=
F
i
⋅
F
j
‖
F
i
‖
‖
F
j
‖
=
∑
k
=
1
m
F
i
k
×
F
j
k
∑
k
=
1
m
(
F
i
m
)
2
×
∑
k
=
1
m
(
F
j
m
)
2
,
where “⋅” denotes the dot product of the two vectors and Fi is the feature vector of case i . The nodule characteristics in the annotated data are used as features for each case, and the corresponding number values of the “gold standard” are normalized and used as the feature values.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Pu,i=∑kl=1[ru,l⋅Sim(i,l)]∑kl=1Sim(i,l), P
u
,
i
=
∑
l
=
1
k
[
r
u
,
l
·
S
i
m
(
i
,
l
)
]
∑
l
=
1
k
S
i
m
(
i
,
l
)
,
where P__u,i is the predication for case i for trainee u , k is the count of the nearest neighbors of case i, and r__u,l is the rating of case l of trainee u . Here, we use a threshold of 0.6 to select the nearest neighbors. For an unrated case in the target trainee vector, if its similar cases are not found in the ratings of the target trainee based on the threshold, then the average rating of the target trainee over the total number of cases becomes its prediction value.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Vu,i={ru,iPu,iiftraineeuratedcaseiotherwise V
u
,
i
=
{
r
u
,
i
i
f
t
r
a
i
n
e
e
u
r
a
t
e
d
c
a
s
e
i
P
u
,
i
o
t
h
e
r
w
i
s
e
Get Radiology Tree app to read full this article<
Step 2: making final prediction using CF algorithm
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Pa,u=∑mi=1(va,i−va¯¯¯)×(vu,i−vu¯¯¯¯)∑mi=1(va,i−va¯¯¯)2×∑mi=1(vu,i−vu¯¯¯¯)2√, P
a
,
u
=
∑
i
=
1
m
(
v
a
,
i
−
v
a
¯
)
×
(
v
u
,
i
−
v
u
¯
)
∑
i
=
1
m
(
v
a
,
i
−
v
a
¯
)
2
×
∑
i
=
1
m
(
v
u
,
i
−
v
u
¯
)
2
,
where m is the number of cases, vu¯¯¯ v
u
¯ is the average rating given by trainee u , and v__a,i is the rating of case i in the pseudotrainee ratings matrix (it is the original rating given by trainee a or a prediction value provided by the CBF predictor).
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Pa,i=va¯¯¯+∑ku=1(vu,i−vu¯¯¯¯)×Pa,u∑ku=1Pa,u, P
a
,
i
=
v
a
¯
+
∑
u
=
1
k
(
v
u
,
i
−
v
u
¯
)
×
P
a
,
u
∑
u
=
1
k
P
a
,
u
,
where P__a,i is the prediction for case i for trainee a , P__a,u is the similarity between trainee a and u , and k is the number of trainees in the neighborhood. For our experiment, we use a threshold of 0.6 to select neighbors.
Get Radiology Tree app to read full this article<
Training Tools
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Results and System Evaluation
Implementation of the Training System
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Experimental Evaluation
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Acknowledgments
Get Radiology Tree app to read full this article<
References
1. Siegel R., Naishadham D., Jemal A.: Cancer statistics, 2013. CA: a cancer journal for clinicians 2013; 63: pp. 11-30.
2. Henschke C.I., McCauley D.I., Yankelevitz D.F., et. al.: Early Lung Cancer Action Project: overall design and findings from baseline screening. The Lancet 1999; 354: pp. 99-105.
3. Schilham A.M.R., Van Ginneken B., Loog M.: A computer-aided diagnosis system for detection of lung nodules in chest radiographs with an evaluation on a public database. Medical Image Analysis 2006; 10: pp. 247-258.
4. Retico A., Delogu P., Fantacci M.E., et. al.: Lung nodule detection in low-dose and thin-slice computed tomography. Computers in Biology and Medicine 2008; 38: pp. 525-534.
5. Hardie R.C., Rogers S.K., Wilson T., et. al.: Performance analysis of a new computer aided detection system for identifying lung nodules on chest radiographs. Medical Image Analysis 2008; 12: pp. 240-258.
6. Messay T., Hardie R.C., Rogers S.K.: A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Medical Image Analysis 2010; 14: pp. 390-406.
7. Reeves A.P., Kostis W.J.: Computer-aided diagnosis for lung cancer. Radiologic Clinics of North America 2000; 38: pp. 497-509.
8. Iwano S., Nakamura T., Kamioka Y., et. al.: Computer-aided diagnosis: a shape classification of pulmonary nodules imaged by high-resolution CT. Computerized Medical Imaging and Graphics 2005; 29: pp. 565-570.
9. Way T.W., Hadjiiski L.M., Sahiner B., et. al.: Computer-aided diagnosis of pulmonary nodules on CT scans: segmentation and classification using 3D active contours. Medical Physics 2006; 33: pp. 2323.
10. Clarke L.P., Croft B.Y., Staab E., et. al.: National Cancer Institute initiative: lung image database resource for imaging research. Academic Radiology 2001; 8: pp. 447-450.
11. McNitt-Gray M.F., Armato S.G., Meyer C.R., et. al.: The Lung Image Database Consortium (LIDC) data collection process for nodule detection and annotation. Academic radiology 2007; 14: pp. 1464-1474.
12. Armato S.G., Roberts R.Y., McNitt-Gray M.F., et. al.: The lung image database consortium (LIDC): ensuring the integrity of expert-defined “truth”. Academic radiology 2007; 14: pp. 1455-1463.
13. Armato S.G., MacMahon H., Engelmann R.M., et. al.: The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Medical physics 2011; 38: pp. 915.
14. Lam M.O., Lam M.O., Disney T., et. al.: BRISC—an open source pulmonary nodule image retrieval framework. Journal of digital imaging 2007; 20: pp. 63-71.
15. Dasovich G.M., Dasovich G., Kim R., et. al.: A model for the relationship between semantic and content based similarity using LIDC. SPIE Medical Imaging. International Society for Optics and Photonics 2010;
16. Zinovev, D., D. Raicu, and J. Furst. Semi-supervised learning approaches for predicting lung nodules semantic characteristics. Margin 35.98.68: 96–83.
17. Armato S.G., McNitt-Gray M.F., Reeves A.P., et. al.: The Lung Image Database Consortium (LIDC): an evaluation of radiologist variability in the identification of lung nodules on CT scans. Academic radiology 2007; 14: pp. 1409-1421.
18. Suzuki K., Shiraishi J., Abe H., et. al.: False-positive reduction in computer-aided diagnostic scheme for detecting nodules in chest radiographs by means of massive training artificial neural network. Academic Radiology 2005; 12: pp. 191-201.
19. Leung J.W., Margolin F.R., Dee K.E., et. al.: Performance parameters for screening and diagnostic mammography in a community practice: are there differences between specialists and general radiologists?. American Journal of Roentgenology 2007; 188: pp. 236-241.
20. Neri E., Faggioni L., Regge D., et. al.: CT colonography: role of a second reader CAD paradigm in the initial training of radiologists. European journal of radiology 2011; 80: pp. 303-309.
21. Taylor P.M.: A review of research into the development of radiologic expertise: implications for computer-based training. Academic radiology 2007; 14: pp. 1252-1263.
22. Guliato D., Ricardo S.B., Marcelo A.M., et. al.: INDIAM—an e-learning system for the interpretation of mammograms. Journal of digital imaging 2009; 22: pp. 405-420.
23. Wu M., Zhang X., Koening L., et. al.: Web-based training method for interpretation of dental images. Journal of Digital Imaging 2010; 23: pp. 493-500.
24. Mazurowski M.A., Baker J.A., Barnhart H.X., et. al.: Individualized computer-aided education in mammography based on user modeling: concept and preliminary experiments. Medical physics 2010; 37: pp. 1152-1160.
25. Mazurowski M.A., Tourassi G.D.: Exploring the potential of collaborative filtering for user-adaptive mammography education. Biomedical Sciences and Engineering Conference (BSEC), 2011. IEEE 2011;
26. Lin H., Yang X., Wang W.: A content-boosted collaborative filtering algorithm for personalized training in interpretation of radiological imaging. Journal of digital imaging 2014; pp. 1-8.
27. Guliato D., de Melo E.V., Rangayyan R.M., et. al.: PostgreSQL-IE: an image-handling extension for PostgreSQL. Journal of digital imaging 2009; 22: pp. 149-165.
28. Herlocker J.L., Konstan J.A., Terveen L.G., et. al.: Evaluating collaborative filtering recommender systems. ACM Transactions on Information Systems (TOIS) 2004; 22: pp. 5-53.
29. Balabanović M., Shoham Y.: Fab: content-based, collaborative recommendation. Communications of the ACM 1997; 40: pp. 66-72.
30. Huang Z., Chen H., Zeng D.: Applying associative retrieval techniques to alleviate the sparsity problem in collaborative filtering. ACM Transactions on Information Systems (TOIS) 2004; 22: pp. 116-142.
31. Linden G., Smith B., York Jeremy: Amazon.com recommendations: Item-to-item collaborative filtering. Internet Computing, IEEE 2003; 7: pp. 76-80.
32. Melville P., Mooney R.J., Nagarajan R.: Content-boosted collaborative filtering for improved recommendations. AAAI/IAAI 2002;
33. Herlocker, J.L., J.A. Konstan, and John Riedl. Explaining collaborative filtering recommendations. Proceedings of the 2000 ACM conference on Computer supported cooperative work. ACM, 2000.