Rationale and Objectives
The purpose of this study was to evaluate the feasibility and technical quality of a zoomed three-dimensional (3D) turbo spin-echo (TSE) sampling perfection with application optimized contrasts using different flip-angle evolutions (SPACE) sequence of the lumbar spine.
Materials and Methods
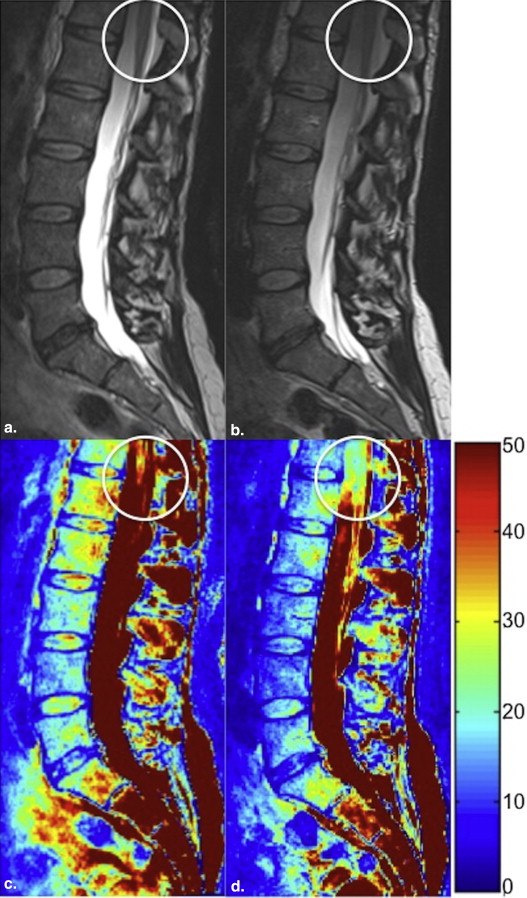
In this prospective feasibility study, nine volunteers underwent a 3-T magnetic resonance examination of the lumbar spine including 1) a conventional 3D T2-weighted (T2w) SPACE sequence with generalized autocalibrating partially parallel acquisition technique acceleration factor 2 and 2) a zoomed 3D T2w SPACE sequence with a reduced field of view (reduction factor 2). Images were evaluated with regard to image sharpness, signal homogeneity, and the presence of artifacts by two experienced radiologists. For quantitative analysis, signal-to-noise ratio (SNR) values were calculated.
Results
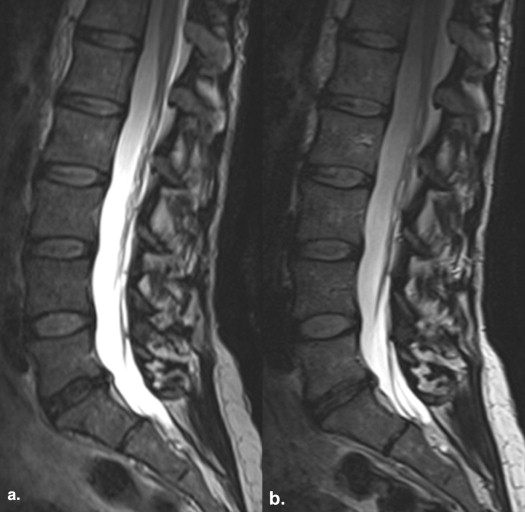
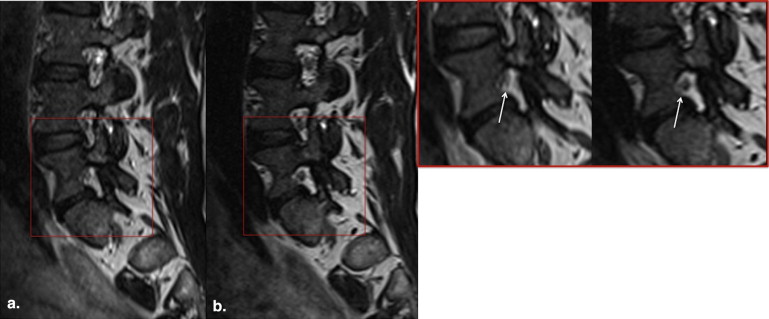
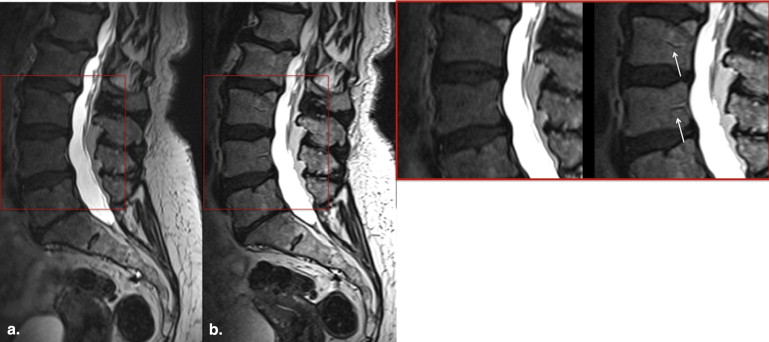
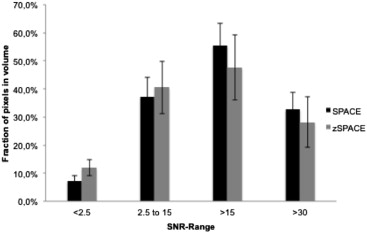
Image sharpness of anatomic structures was statistically significantly greater with zoomed SPACE ( P < .0001), whereas the signal homogeneity was statistically significantly greater with conventional SPACE (cSPACE; P = .0003). There were no statistically significant differences in extent of artifacts. Acquisition times were 8:20 minutes for cSPACE and 6:30 minutes for zoomed SPACE. Readers 1 and 2 selected zSPACE as the preferred sequence in five of nine cases. In two of nine cases, both sequences were rated as equally preferred by both the readers. SNR values were statistically significantly greater with cSPACE.
Conclusions
In comparison to a cSPACE sequences, zoomed SPACE imaging of the lumbar spine provides sharper images in conjunction with a 25% reduction in acquisition time.
Magnetic resonance imaging (MRI) is the modality of choice for evaluation of the spine and degenerative disc disease because of its excellent soft tissue contrast. T2-weighted (T2W), two-dimensional (2D), turbo spin-echo (TSE) sequences provide high contrast between anatomic structures enabling reliable visualization of spinal lesions and pathologic changes within the intervertebral discs . Nevertheless, with 2D sequences, through-plane resolution is limited because of signal-to-noise ratio (SNR) loss, increases in examination time, and cross-talk artifact. The latter further reduces SNR and alters image contrast .
Conventional, fast 3D imaging techniques are primarily based on gradient-echo pulse sequences that result in T1 or T2* contrast. However, adequate T2 contrast is required for the diagnosis of degenerative spine disorders, a condition present in a large proportion of examinations performed in the daily clinical routine . Although adequate image contrast is achievable with gradient-echo magnetic resonance (MR) sequences (eg, multi echo data image combination (MEDIC) or double-echo steady state (DESS) sequences) , a spin-echo or TSE sequence provides optimal T2 contrast. At 3 T, spin-echo–based techniques are limited by high specific absorption rates (SARs). Such SAR considerations have encouraged the development of a three-dimensional (3D), T2w, TSE sequence with variable flip angles along the echo train (sampling perfection with application optimized contrasts using different flip-angle evolutions [SPACE]) . Three-dimensional sequences with isotropic spatial resolution enable high-quality reformatted images in arbitrary spatial orientations, potentially improving diagnostic performance. Three-dimensional T2w SPACE has been shown to be clinically feasible for high-resolution imaging of the cervical spine and superior to conventional 2D T2w TSE imaging in the delineation of anatomic details in that region .
Get Radiology Tree app to read full this article<
Materials and methods
Get Radiology Tree app to read full this article<
Image Acquisition
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 1
Imaging Parameters for the Volunteer and Patient Populations
Sequence Type cSPACE zSPACE 3D SPACE TR/TE (ms) 1700/138 1700/138 FOV (mm 2 ) 300 × 300 150 × 320 Matrix 304 × 320 160 × 320 Slice thickness (mm) 0.9 0.9 In-plane resolution (mm 2 ) 0.9 × 0.9 0.9 × 0.9 Phase-encoding direction Head–feet Anterior–posterior Phase-encoding steps 433 166 Parallel imaging GRAPPA2 — Acquisition time (min) 8:20 6:30 Averages 2 2 Flip angle 100° 100° Bandwidth (Hz/px) 380 380
3D, three-dimensional; cSPACE, conventional SPACE; FOV, field of view; GRAPPA, generalized autocalibrating partially parallel acquisition technique; SPACE, sampling perfection with application optimized contrasts using different flip-angle evolutions; TE, echo time; TR, repetition time; zSPACE, zoomed SPACE.
Get Radiology Tree app to read full this article<
Image Analysis
Qualitative Image analysis
Get Radiology Tree app to read full this article<
Quantitative Image Analysis
Get Radiology Tree app to read full this article<
Statistical Analysis
Get Radiology Tree app to read full this article<
Results
Qualitative Image Analysis
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 2
Image Quality Scores (Median) of Both Sequences
Image quality parameters cSPACE zSPACE Sharpness 2 3 Image homogeneity 3 2 Artifacts 3 3
cSPACE, conventional SPACE; SPACE, sampling perfection with application optimized contrasts using different flip-angle evolutions; zSPACE, zoomed SPACE.
Get Radiology Tree app to read full this article<
Quantitative Image Analysis
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
References
1. Meindl T., Wirth S., Weckbach S., et. al.: Magnetic resonance imaging of the cervical spine: comparison of 2D T2-weighted turbo spin echo, 2D T2*weighted gradient-recalled echo and 3D T2-weighted variable flip-angle turbo spin echo sequences. Eur Radiol 2009; 19: pp. 713-721.
2. Boutin R.D., Steinbach L.S., Finnesey K.: MR imaging of degenerative diseases in the cervical spine. Magn Reson Imaging Clin N Am 2000; 8: pp. 471-490.
3. Melhem E.R.: Technical challenges in MR imaging of the cervical spine and cord. Magn Reson Imaging Clin N Am 2000; 8: pp. 435-452.
4. Hayes C.W., Jensen M.E., Conway W.F.: Non-neoplastic lesions of vertebral bodies: findings in magnetic resonance imaging. Radiographics 1989; 9: pp. 883-903.
5. Freund M., Sartor K.: Degenerative spine disorders in the context of clinical findings. Eur J Radiol 2006; 58: pp. 15-26.
6. Held P., Dorenbeck U., Seitz J., et. al.: MRI of the abnormal cervical spinal cord using 2D spoiled gradient echo multiecho sequence (MEDIC) with magnetization transfer saturation pulse. A T2* weighted feasibility study. J Neuroradiol 2003; 30: pp. 83-90.
7. Muhle C., Ahn J.M., Biederer J., et. al.: MR imaging of the neural foramina of the cervical spine. Comparison of 3D-DESS and 3D-FISP sequences. Acta Radiol 2002; 43: pp. 96-100.
8. Lichy M.P., Wietek B.M., Mugler J.P., et. al.: Magnetic resonance imaging of the body trunk using a single-slab, 3-dimensional, T2-weighted turbo-spin-echo sequence with high sampling efficiency (SPACE) for high spatial resolution imaging: initial clinical experiences. Invest Radiol 2005; 40: pp. 754-760.
9. Mugler J.P., Bao S., Mulkern R.V., et. al.: Optimized single-slab three-dimensional spin-echo MR imaging of the brain. Radiology 2000; 216: pp. 891-899.
10. Feinberg D.A., Hoenninger J.C., Crooks L.E., et. al.: Inner volume MR imaging: technical concepts and their application. Radiology 1985; 156: pp. 743-747.
11. Mitsouras D., Mulkern R.V., Rybicki F.J.: Strategies for inner volume 3D fast spin echo magnetic resonance imaging using nonselective refocusing radio frequency pulses. Med Phys 2006; 33: pp. 173-186.
12. Robson P.M., Grant A.K., Madhuranthakam A.J., et. al.: Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn Reson Med 2008; 60: pp. 895-907.
13. Wiens C.N., Kisch S.J., Willig-Onwuachi J.D., et. al.: Computationally rapid method of estimating signal-to-noise ratio for phased array image reconstructions. Magn Reson Med 2011; 66: pp. 1192-1197.
14. Stevens K.J., Busse R.F., Han E., et. al.: Ankle: isotropic MR imaging with 3D-FSE-cube—initial experience in healthy volunteers. Radiology 2008; 249: pp. 1026-1033.
15. Kwon J.W., Yoon Y.C., Choi S.H.: Three-dimensional isotropic T2-weighted cervical MRI at 3T: comparison with two-dimensional T2-weighted sequences. Clin Radiol 2012; 67: pp. 106-113.
16. Alley M.T., Pauly J.M., Sommer F.G., et. al.: Angiographic imaging with 2D RF pulses. Magn Reson Med 1997; 37: pp. 260-267.
17. Yang G.Z., Burger P., Gatehouse P.D., et. al.: Locally focused 3D coronary imaging using volume-selective RF excitation. Magn Reson Med 1999; 41: pp. 171-178.
18. Luk-Pat G.T., Gold G.E., Olcott E.W., et. al.: High-resolution three-dimensional in vivo imaging of atherosclerotic plaque. Magn Reson Med 1999; 42: pp. 762-771.
19. Botnar R.M., Kim W.Y., Bornert P., et. al.: 3D coronary vessel wall imaging utilizing a local inversion technique with spiral image acquisition. Magn Reson Med 2001; 46: pp. 848-854.
20. Maier S.E.: Slab scan diffusion imaging. Magn Reson Med 2001; 46: pp. 1136-1143.
21. Rieseberg S., Frahm J., Finsterbusch J.: Two-dimensional spatially-selective RF excitation pulses in echo-planar imaging. Magn Reson Med 2002; 47: pp. 1186-1193.
22. Crema M.D., Roemer F.W., Marra M.D., et. al.: Articular cartilage in the knee: current MR imaging techniques and applications in clinical practice and research. Radiographics 2011; 31: pp. 37-61.