Rationale and Objectives
Accuracy and speed are essential for the intraprocedural nonrigid magnetic resonance (MR) to computed tomography (CT) image registration in the assessment of tumor margins during CT-guided liver tumor ablations. Although both accuracy and speed can be improved by limiting the registration to a region of interest (ROI), manual contouring of the ROI prolongs the registration process substantially. To achieve accurate and fast registration without the use of an ROI, we combined a nonrigid registration technique on the basis of volume subdivision with hardware acceleration using a graphics processing unit (GPU). We compared the registration accuracy and processing time of GPU-accelerated volume subdivision–based nonrigid registration technique to the conventional nonrigid B-spline registration technique.
Materials and Methods
Fourteen image data sets of preprocedural MR and intraprocedural CT images for percutaneous CT-guided liver tumor ablations were obtained. Each set of images was registered using the GPU-accelerated volume subdivision technique and the B-spline technique. Manual contouring of ROI was used only for the B-spline technique. Registration accuracies (Dice similarity coefficient [DSC] and 95% Hausdorff distance [HD]) and total processing time including contouring of ROIs and computation were compared using a paired Student t test.
Results
Accuracies of the GPU-accelerated registrations and B-spline registrations, respectively, were 88.3 ± 3.7% versus 89.3 ± 4.9% ( P = .41) for DSC and 13.1 ± 5.2 versus 11.4 ± 6.3 mm ( P = .15) for HD. Total processing time of the GPU-accelerated registration and B-spline registration techniques was 88 ± 14 versus 557 ± 116 seconds ( P < .000000002), respectively; there was no significant difference in computation time despite the difference in the complexity of the algorithms ( P = .71).
Conclusions
The GPU-accelerated volume subdivision technique was as accurate as the B-spline technique and required significantly less processing time. The GPU-accelerated volume subdivision technique may enable the implementation of nonrigid registration into routine clinical practice.
Computed tomography (CT) is used commonly to guide percutaneous tumor ablations of liver tumors . CT can provide high-quality three-dimensional (3D) images of the liver intraprocedurally, which help radiologists understand spatial relationship of the tumor with respect to the surrounding structures . However, typically, only sectional unenhanced CT images are used, and hence, tumor margins may not be delineated well ; this limitation may contribute to inadequate ablation . A recent study showed that the rate of local recurrence following percutaneous radiofrequency ablation of hepatocellular carcinoma decreased significantly from 23% to 0% when the ablation margin, the shortest distance between the outer margin of the tumor and the outer margin of the ablation, was increased from <1 to >3 mm . Therefore, visualizing tumor margins intraprocedurally is important. One way to delineate tumor margins intraprocedurally is to register preprocedural magnetic resonance (MR) images to the intraprocedural CT images; tumor margins depicted by MRI can be directly compared to ablation effects depicted with intraprocedural CT . In addition, the same data can be used to depict tumor and ablation volumes .
Nonrigid image registration techniques can be used to correct accurately the misalignment between structures in two images caused by physiologic variations . For example, the liver may be misaligned because of diaphragmatic motion. B-spline registration is a commonly used nonrigid registration technique that parameterizes a deformation of an image as displacements of control points on a regular grid and calculates the displacements of the voxels between the control points using basis spline (B-spline) interpolation . However, nonrigid registration techniques are not widely used clinically for intraprocedural registration because the registration takes too long to perform and often needs a dedicated software operator. Although computation time for B-spline registration can be shortened by reducing the control points, fewer control points may impact the registration accuracy negatively. In particular, the B-spline cannot model local deformations well with limited control points. A previous analysis of four-dimensional CT by Schreibmann et al. used 15 control points per direction to model the deformations of liver images accurately. This number of control points is much larger than those used in previous studies on intraoperative registration, typically 3–5 control points per direction, which still takes up to 2 minutes . To attain sufficient registration accuracy, a region of interest (ROI) is often used . An ROI can restrict the region to be registered on the liver only and can reduce the effect of local displacement in the surrounding structures. An ROI also helps reduce computation time by reducing the number of voxels to be processed. However, an ROI must be defined by contouring the target organ. Robust automatic contouring of the target organ is a challenging problem and would invariably introduce additional computation time. Manual contouring has been used in past studies but would require an additional human resource for routine clinical use. The long computation time and the need for manual contouring hinder widespread use of nonrigid image registration.
Get Radiology Tree app to read full this article<
Materials and methods
Subjects
Get Radiology Tree app to read full this article<
Imaging Protocol
Get Radiology Tree app to read full this article<
Image Registration Algorithms
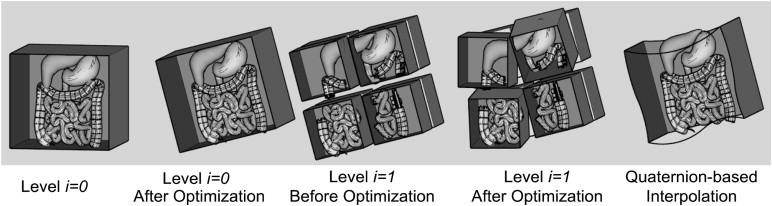
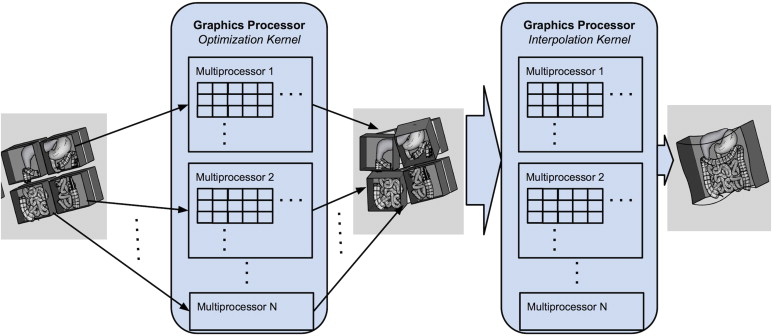
GPU-Accelerated Nonrigid Registration Using Volume Subdivision Approach
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Nonrigid B-Spline Registration
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Comparison of Registration Accuracy and Processing Time
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
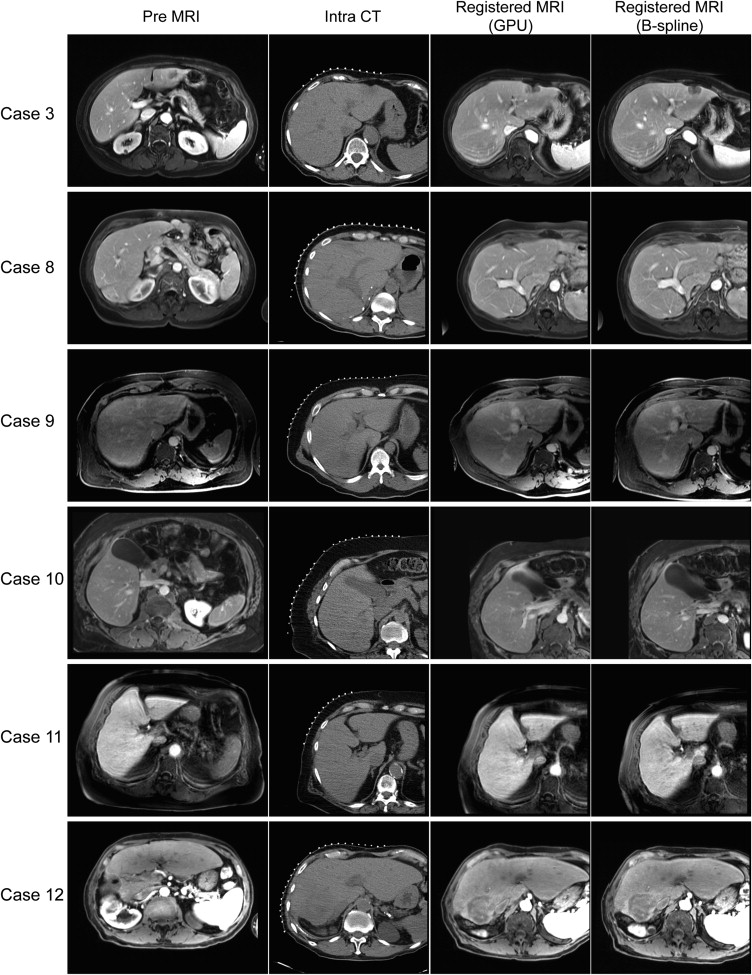
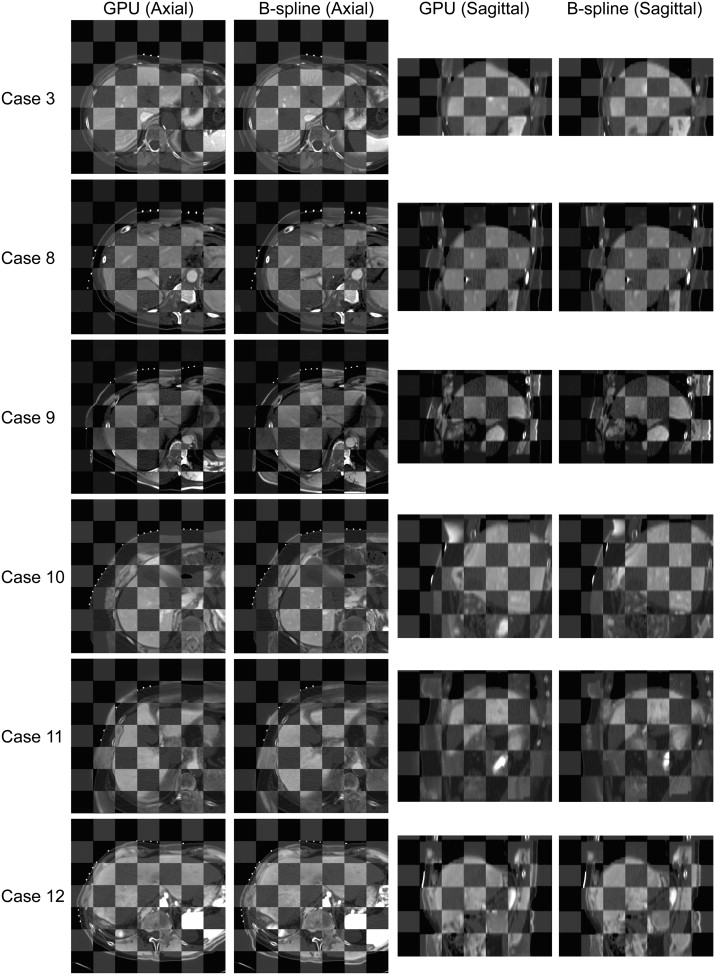
Results
Get Radiology Tree app to read full this article<
Table 1
Summary of Results of Nonrigid Registration Between Preprocedural MRI and Intraprocedural CT Images Using B-Spline and GPU-Accelerated Registration Techniques
Mean ± SD Range_P_ Value DSC (%) .41 GPU 88.30 ± 3.74 80.49–93.11 B-Spline 89.32 ± 4.90 79.70–94.46 95% Hausdorff distance (mm) .15 GPU 13.14 ± 5.19 6.43–24.54 B-Spline 11.44 ± 6.32 5.43–23.04 Time, rigid reg. (s) <.002 GPU 8.58 ± 6.31 4.36–29.28 B-Spline 20.53 ± 7.01 14.56–40.08 Time, nonrigid reg. (s) <.00002 GPU 76.90 ± 10.75 61.5–91.43 B-Spline 42.48 ± 11.39 19.38–68.57 Time, resampling (s) <.0000000001 GPU 2.07 ± 0.47 1–3 B-Spline 27.72 ± 5.02 19.87–40.71 Time, total computation (s) .71 GPU 87.55 ± 14.26 69.10–118.25 B-Spline 90.72 ± 21.81 58.42–149.36 Voxel throughput (voxels/s) <.0004 GPU 195024.67 ± 141785.32 104192.50–614978.60 B-Spline 18,050.36 ± 6324.58 12001.98–32628.07 Time, manual ROI contouring NA GPU NA NA B-Spline 466 ± 104 300–701 Time, total processing <.000000002 GPU 87.55 ± 14.26 69.10–118.25 B-Spline 557.08 ± 116.47 375.83–850.36
CT, computed tomography; GPU, graphics processing unit; MRI, magnetic resonance imaging; NA, not applicable; ROI, region of interest.
Table 2
Type of MRI Scanner, Phase of CE MRI Used for Registration, and Registration Performances Including DSC, 95% HD, Processing Time, and Voxel Throughput for Each Subject
Case MRI Scanner (T) CE MRI Phase DSC (%) HD (mm) Process Time: Rigid Reg. (s) Process Time: Nonrigid Reg. (s) Process Time: Resampling (s) Total Process Time (s) Time: Manual Contouring (s) Total Workflow Time (s) Voxel Throughput (Voxels/s) GPU BS GPU BS GPU BS GPU BS GPU BS GPU BS GPU BS GPU BS GPU BS 1 1.5 2 87.9 87.7 13.1 18.1 7.0 40.1 65.3 68.6 2.0 40.7 74.3 149.4 — 701.0 74.3 850.4 614978.6 29900.7 2 3.0 2 83.0 79.7 24.5 23.0 9.7 16.0 90.6 36.6 2.0 29.4 102.4 82.1 — 499.0 102.4 581.1 107551.5 22465.2 3 3.0 3 91.7 92.4 10.0 10.2 5.2 23.7 62.9 51.9 1.0 28.9 69.1 104.6 — 419.0 69.1 523.6 288972.5 18045.4 4 1.5 1 88.5 93.4 10.2 5.6 4.9 26.5 81.5 51.4 2.0 33.7 88.4 111.6 — 512.0 88.4 623.6 237207.6 12002.0 5 3.0 1 80.5 88.7 20.9 9.0 7.3 16.0 83.8 38.9 2.0 24.1 93.1 79.0 — 550.0 93.1 629.0 109813.3 14060.3 6 3.0 2 90.3 80.5 12.0 19.7 12.0 14.6 72.1 34.4 2.0 24.7 86.1 73.7 — 484.0 86.1 557.7 106612.1 17557.9 7 1.5 3 84.2 83.1 18.7 13.5 4.4 26.9 65.5 42.4 2.0 19.9 71.9 89.2 — 571.0 71.9 660.2 335427.6 17322.2 8 1.5 2 93.1 93.2 6.4 6.6 9.0 21.2 61.5 46.5 2.0 27.8 72.5 95.5 — 417.0 72.5 512.5 190781.1 14972.4 9 3.0 3 89.2 91.0 10.0 6.4 5.2 19.3 74.4 43.0 2.0 26.1 81.6 88.4 — 420.0 81.6 508.4 135680.3 13036.0 10 3.0 3 89.1 89.6 13.2 20.4 29.3 15.1 87.0 19.4 2.0 23.9 118.3 58.4 — 518.0 118.3 576.4 104192.5 32628.1 11 3.0 3 85.1 92.8 16.4 6.1 7.0 19.6 91.4 51.1 3.0 30.5 101.4 101.2 — 429.0 101.4 530.2 131795.6 12035.0 12 3.0 1 91.1 90.6 10.8 10.2 7.3 15.7 90.1 36.0 3.0 26.7 100.3 78.4 — 371.0 100.3 449.4 122826.9 18354.8 13 3.0 2 90.5 93.3 9.3 6.0 5.8 14.8 71.4 36.1 2.0 25.0 79.2 75.8 — 300.0 79.2 375.8 109240.5 13329.1 14 1.5 2 91.9 94.5 8.4 5.4 6.2 18.1 79.0 38.2 2.0 26.7 87.2 83.0 — 338.0 87.2 421.0 135265.2 16996.0
BS, B-Spline; CE, contrast-enhanced; DSC, Dice similarity coefficient; GPU, graphics processing unit; HD, Hausdorff distance; MRI, magnetic resonance imaging.
CE MRI phase: 1 = 30 s; 2 = 60 s; 3 = 90s.
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Orlacchio A., Bazzocchi G., Pastorelli D., et. al.: Percutaneous cryoablation of small hepatocellular carcinoma with US guidance and CT monitoring: initial experience. Cardiovasc. Intervent. Radiol 2008; 31: pp. 587-594.
2. Antoch G., Kuehl H., Vogt F.M., et. al.: Value of CT volume imaging for optimal placement of radiofrequency ablation probes in liver lesions. J. Vasc. Interv. Radiol 2002 Nov; 13: pp. 1155-1161.
3. Sato M., Watanabe Y., Tokui K., et. al.: CT-guided treatment of ultrasonically invisible hepatocellular carcinoma. Am. J. Gastroenterol 2000; 95: pp. 2102-2106.
4. Goshima S., Kanematsu M., Kondo H., et. al.: MDCT of the liver and hypervascular hepatocellular carcinomas: optimizing scan delays for bolus-tracking techniques of hepatic arterial and portal venous phases. AJR. Am. J. Roentgenol 2006; 187: pp. W25-W32.
5. Paushter D.M., Zeman R.K., Scheibler M.L., et. al.: CT evaluation of suspected hepatic metastases: comparison of techniques for i.v. contrast enhancement. AJR. Am. J. Roentgenol 1989; 152: pp. 267-271.
6. Montorsi M., Santambrogio R., Bianchi P., et. al.: Survival and recurrences after hepatic resection or radiofrequency for hepatocellular carcinoma in cirrhotic patients: a multivariate analysis. J Gastrointest Surg 2005; 9: pp. 62-68.
7. Chen M.S., Li J.Q., Zheng Y., et. al.: A prospective randomized trial comparing percutaneous local ablative therapy and partial hepatectomy for small hepatocellular carcinoma. Ann Surg 2006; 243: pp. 321-328.
8. Abu-Hilal M., Primrose J.N., Casaril A., et. al.: Surgical resection versus radiofrequency ablation in the treatment of small unifocal hepatocellular carcinoma. J Gastrointest Surg 2008; 12: pp. 1521-1526.
9. Liang H.H., Chen M.S., Peng Z.W., et. al.: Percutaneous radiofrequency ablation versus repeat hepatectomy for recurrent hepatocellular carcinoma: a retrospective study. Ann Surg Oncol 2008; 15: pp. 3484-3493.
10. Peng Z.W., Zhang Y.J., Chen M.S., et. al.: Radiofrequency ablation as first-line treatment for small solitary hepatocellular carcinoma: long-term results. Eur J Surg Oncol 2010; 36: pp. 1054-1060.
11. Kim Y.S., Lee W.J., Rhim H., et. al.: The minimal ablative margin of radiofrequency ablation of hepatocellular carcinoma (>2 and <5 cm) needed to prevent local tumor progression: 3D quantitative assessment using CT image fusion. AJR Am J Roentgenol 2010; 195: pp. 758-765.
12. Elhawary H., Oguro S., Tuncali K., et. al.: Multimodality non-rigid image registration for planning, targeting and monitoring during CT-guided percutaneous liver tumor cryoablation. Acad Radiol 2010; 17: pp. 1334-1344. Elsevier Ltd
13. Rohlfing T., Maurer C.R., O’Dell W.G., et. al.: Modeling liver motion and deformation during the respiratory cycle using intensity-based nonrigid registration of gated MR images. Med Phys 2004; 31: pp. 427-432.
14. Rueckert D., Sonoda L.I., Hayes C., et. al.: Nonrigid registration using free-form deformations: application to breast MR images. Med. Imaging, IEEE Trans 1999; 18: pp. 712-721.
15. Wells W.M., Viola P., Atsumi H., et. al.: Multi-modal volume registration by maximization of mutual information. Med Image Anal 1996; 1: pp. 35-51.
16. Fedorov A., Tuncali K., Fennessy F.M., et. al.: Image registration for targeted MRI-guided transperineal prostate biopsy. J Magn Reson Imaging 2012; 36: pp. 987-992.
17. Schreibmann E., Chen G.T.Y., Xing L.: Image interpolation in 4D CT using a BSpline deformable registration model. Int. J. Radiat. Oncol. Biol. Phys 2006; 64: pp. 1537-1550.
18. Rietzel E., Chen G.T.Y.: Deformable registration of 4D computed tomography data. Med. Phys. American Association of Physicists in Medicine 2006; 33: pp. 4423.
19. Oguro S., Tokuda J., Elhawary H., et. al.: MRI signal intensity based B-spline nonrigid registration for pre- and intraoperative imaging during prostate brachytherapy. J. Magn. Reson. Imaging 2009; 30: pp. 1052-1058.
20. Hellier P., Barillot C., Mémin E., et. al.: Hierarchical estimation of a dense deformation field for 3-D robust registration. IEEE Trans Med Imaging 2001; 20: pp. 388-402.
21. Walimbe V., Shekhar R.: Automatic elastic image registration by interpolation of 3D rotations and translations from discrete rigid-body transformations. Med. Image Anal 2006; 10: pp. 899-914.
22. Klein S., Staring M., Murphy K., et. al.: Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010; 29: pp. 196-205.
23. Shams R., Sadeghi P., Kennedy R., et. al.: A survey of medical image registration on multicore and the GPU. IEEE Signal Process. Mag. IEEE 2010; 27: pp. 50-60.
24. Ito K., Choji T., Nakada T., et. al.: Multislice dynamic MRI of hepatic tumors. J Comput Assist Tomogr 1993; 17: pp. 390-396.
25. Yamashita Y., Mitsuzaki K., Yi T., et. al.: Small hepatocellular carcinoma in patients with chronic liver damage: prospective comparison of detection with dynamic MR imaging and helical CT of the whole liver. Radiology 1996; 200: pp. 79-84.
26. Peterson M.S., Baron R.L., Murakami T.: Hepatic malignancies: usefulness of acquisition of multiple arterial and portal venous phase images at dynamic gadolinium-enhanced MR imaging. Radiology 1996; 201: pp. 337-345.
27. Semelka R.C., Martin D.R., Balci C., et. al.: Focal liver lesions: comparison of dual-phase CT and multisequence multiplanar MR imaging including dynamic gadolinium enhancement. J Magn Reson Imaging 2001; 13: pp. 397-401.
28. Gering D.T., Nabavi A., Kikinis R., et. al.: An integrated visualization system for surgical planning and guidance using image fusion and an open MR. J Magn Reson Imaging 2001; 13: pp. 967-975.
29. Fedorov A., Beichel R., Kalpathy-Cramer J., et. al.: 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012; 30: pp. 1323-1341.
30. Studholme C., Hill D.L.G., Hawkes D.J.: An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognit 1999; 32: pp. 71-86.
31. Shoemake K.: Quaternion calculus and fast animation, computer animation: 3-D motion specification and control.1987.SIGGRAPH’87
32. Tustison N.J., Avants B.B., Cook P.A., et. al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 2010; 29: pp. 1310-1320.
33. Arnold J.B., Liow J.S., Schaper K.A., et. al.: Qualitative and quantitative evaluation of six algorithms for correcting intensity nonuniformity effects. Neuroimage 2001; 13: pp. 931-943.
34. Boyes R.G., Gunter J.L., Frost C., et. al.: Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. Neuroimage 2008; 39: pp. 1752-1762.
35. Zheng W., Chee M.W.L., Zagorodnov V.: Improvement of brain segmentation accuracy by optimizing non-uniformity correction using N3. Neuroimage 2009; 48: pp. 73-83.
36. Johnson H.J., Harris G., Williams K.: BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit. Insight J 2007;
37. Mattes D., Haynor D.R., Vesselle H., et. al.: PET-CT image registration in the chest using free-form deformations. Med. Imaging, IEEE Trans 2003; 22: pp. 120-128.
38. Byrd R.H., Lu P., Nocedal J., et. al.: A limited memory algorithm for bound constrained optimization. SIAM J. Sci. Comput 1995; 16: pp. 1190-1208. SIAM PUBLICATIONS, 3600 UNIV CITY SCIENCE CENTER PH#382-9800, PHILADELPHIA, PA 19104-2688
39. Bharatha A., Hirose M., Hata N., et. al.: Evaluation of three-dimensional finite element-based deformable registration of pre- and intraoperative prostate imaging. Med Phys 2001; 28: pp. 2551-2560.
40. Zou K.H., Warfield S.K., Bharatha A., et. al.: Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 2004; 11: pp. 178-189.
41. Morikawa S., Inubushi T., Kurumi Y., et. al.: MR-guided microwave thermocoagulation therapy of liver tumors: initial clinical experiences using a 0.5 T open MR system. J. Magn. Reson. Imaging 2002; 16: pp. 576-583.
42. Morrison P.R., Silverman S.G., Tuncali K., et. al.: MRI-guided cryotherapy. J. Magn. Reson. Imaging 2008; 27: pp. 410-420.
43. Rempp H., Waibel L., Hoffmann R., et. al.: MR-guided radiofrequency ablation using a wide-bore 1.5-T MR system: clinical results of 213 treated liver lesions. Eur. Radiol 2012; 22: pp. 1972-1982.
44. Passera K., Selvaggi S., Scaramuzza D., et. al.: Radiofrequency ablation of liver tumors: quantitative assessment of tumor coverage through CT image processing. BMC Med. Imaging 2013; 13: pp. 3.
45. Asanovic K., Bodik R., Demmel J., et. al.: A view of the parallel computing landscape. Commun. ACM 2009; 52: pp. 56-67.
46. Xie Y., Chao M., Xiong G.: Deformable image registration of liver with consideration of lung sliding motion. Med. Phys 2011; 38: pp. 5351-5361.