Recent discoveries characterizing the molecular basis of lung cancer brought fundamental changes in lung cancer treatment. The authors review the molecular pathogenesis of lung cancer, including genomic abnormalities, targeted therapies, and resistance mechanisms, and discuss lung cancer imaging with novel techniques. Knowledge of the molecular basis of lung cancer is essential for radiologists to properly interpret imaging and assess response to therapy. Quantitative and functional imaging helps assessing the biologic behavior of lung cancer.
Lung cancer remains the leading cause of cancer deaths for both men and women in the United States and worldwide, accounting for 30% of estimated cancer deaths in men and 26% of estimated cancer deaths in women in the United States in 2009 ( Fig 1 ) . In addition, the mortality rate of lung cancer is much higher than that of other top three causes of cancer death, including breast, prostate, and colon cancer ( Fig 2 ) . Eighty-five percent of patients with lung cancer have non-small-cell lung cancer (NSCLC), for which the 5-year survival rate is only 15% . Two thirds of patients with NSCLC present with advanced disease and are considered incurable by surgery or radiotherapy. Platinum-based doublet chemotherapy, the standard of care for these patients, is also marginally effective. It has been clear in the past decades that more effective systemic therapy is needed for patients with advanced lung cancer .
Figure 1
Estimated cancer deaths in women in the United States in 2009.
Modified from American Cancer Society .
Figure 2
Incidence and mortality rates for four leading causes of cancer death.
Modified from American Cancer Society .
Recent advances in molecular biology have elucidated the different molecular mechanisms of lung cancer development and progression. Some of these genetic abnormalities are specific to lung cancer, while others are present in other cancers. One of the major discoveries was the identification of somatic activating mutations of the epidermal growth factor receptor (EGFR) tyrosine kinase domain in NSCLC. The somatic mutations of EGFR are associated with a dramatic clinical response to the EGFR tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib . The discovery of EGFR mutations and the clinical application of this finding for the selection of therapy have transformed the way oncologists approach lung cancer and plan treatment. It is also a pragmatic example of the contributions that advances in basic molecular research have made to patient care in clinical oncology. As radiologists involved in imaging of patients with lung cancer, we should be familiar with these molecular bases of determining therapies and applications that our oncology colleagues are using. Radiologists should become familiar with the molecular background of lung cancer and its new molecular-targeted treatment approach, to properly validate, use, and apply our advanced imaging technology to diagnose, assess response, and define progressive disease. This will help radiologists contribute in a clinically significant manner as cancer imaging specialists to the management and further progress of care for patients with lung cancer.
In this review article, we describe different molecular mechanisms of the pathogenesis of lung cancer, initially focusing on EGFR mutations in NSCLC, their therapeutic application, and current challenges. The role of histology in lung cancer assessment in current clinical oncology is also discussed. We present information on different imaging approaches to lung cancer, including conventional response assessment methods and their limitations, newer quantitative and functional imaging with multi–detector row computed tomographic (CT) imaging, dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI), and combined positron emission tomographic (PET) and CT imaging. The current status of these advanced imaging techniques in lung cancer is described, their current challenges are outlined, and future directions are proposed.
Molecular mechanisms of lung cancer
EGFR Mutation and Its Inhibitors in NSCLC
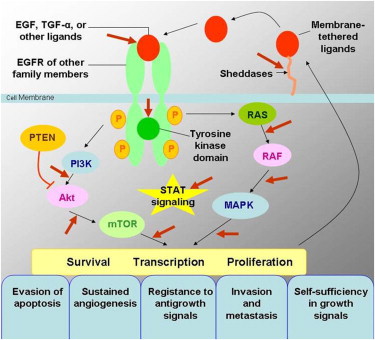
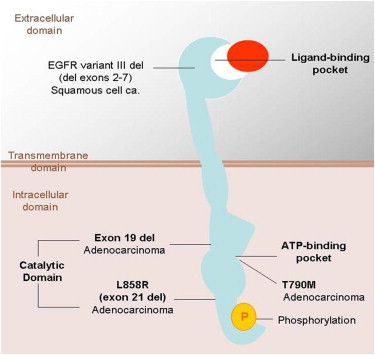
The EGFR is a transmembrane tyrosine kinase receptor involved in signaling pathways of cells, and it regulates important tumorigenic processes, including proliferation, apoptosis, angiogenesis, and invasion ( Fig 3 ) . Overexpression of EGFR is frequently noted in the development and progression of NSCLC, and its presence is associated with shortened survival . To specifically target this EGFR pathway in NSCLC, small-molecule inhibitors of the tyrosine kinase domain of EGFR were developed, and erlotinib and gefitinib have been approved for therapy of patients with advanced NSCLC in different parts of the world . Subsequently, activating EGFR mutations were discovered in cancer cells from patients with NSCLC who responded to the targeted therapy with gefitinib and erlotinib .
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Rat Sarcoma ( RAS ) Mutations in NSCLC
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 1
Predictors of Sensitivity to Erlotinib Therapy in Non-Small-Cell Lung Cancer
Modified from Mol Oncol Rep .
Positive Predictors Clinical Molecular Nonsmoker status_EGFR_ mutations (exon 19 deletions, L858R point mutations) Asian ethnicity_EGFR_ gene amplification (FISH or CISH) Female gender EGFR protein expression on immunohistochemistry Adenocarcinoma/BAC histology MALDI-ToF algorithm
Negative predictors_KRAS_ mutations T790M mutations Exon 20 insertion mutations
BAC, bronchioloalveolar cell carcinoma; CISH, chromogenic in situ hybridization; FISH, fluorescence in situ hybridization; KRAS , Vi-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog; L858R, substitution of arginine for leucine at amino acid position 858; MALDI-ToF, matrix-assisted laser desorption/ionization–time of flight; T790M, substitution of methionine for threonine at amino acid position 790.
Get Radiology Tree app to read full this article<
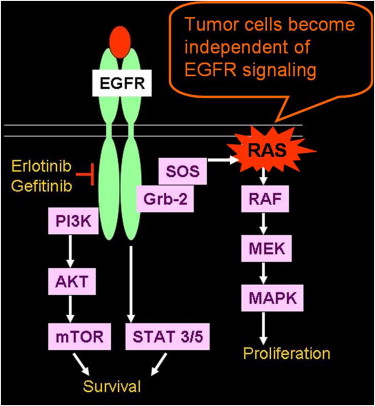
Acquired Resistance to Erlotinib
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Histology and genomic mutation as markers for therapy
Assessment of Lung Cancer in the New Era of Molecular Medicine
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Histology as a Marker for Therapy
Get Radiology Tree app to read full this article<
Table 2
Histology and Genomic Mutations in Lung Cancer
Histology Genomic Mutations Adenocarcinoma (50%)EGFR , KRAS , TP53 , STK11 , CDKN2A , ERBB-2 Squamous cell carcinoma (20%) SOX2 amplification , EGFR VIII Small-cell lung cancer (15%)TP53 , RB1 , Myc gene amplification , nontargetable oncogene
CDKN2A , cyclin-dependent kinase inhibitor 2A; ERBB-2 , erythroblastic leukemia viral oncogene homolog 2; RB1 , retinoblastoma 1; SOX2, sex-determining region Y–box 2; STK11 , serine/threonine kinase 11; TP53 , tumor protein 53.
Get Radiology Tree app to read full this article<
Genomewide Approach to Characterize Lung Cancer Genomes
Get Radiology Tree app to read full this article<
Lung cancer imaging in assessment of response to therapy
Get Radiology Tree app to read full this article<
Response Assessment on the Basis of Size Measurement
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
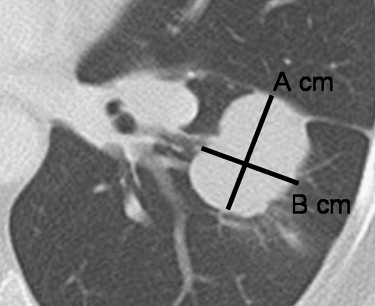
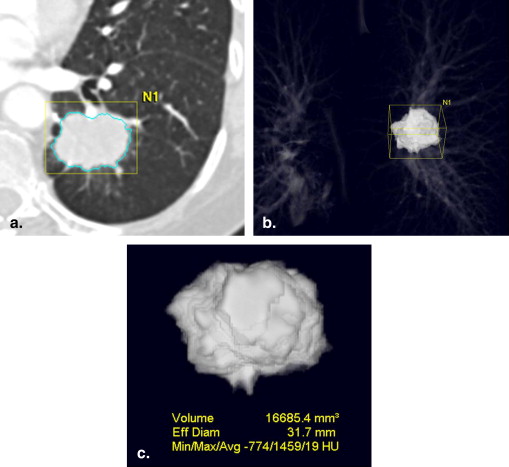
Tumor Volume Measurement Using Multidetector CT Imaging
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
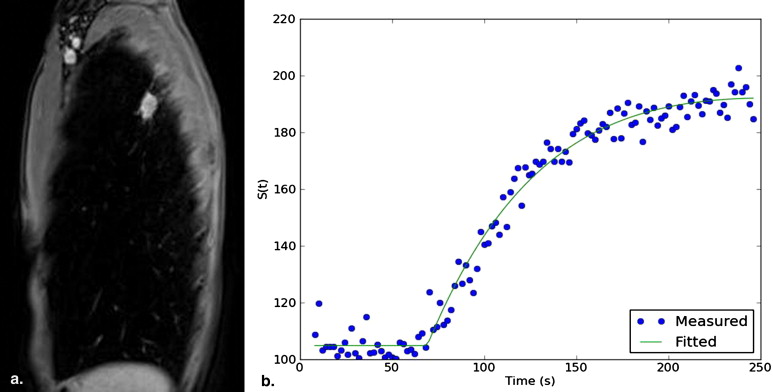
DCE MRI
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
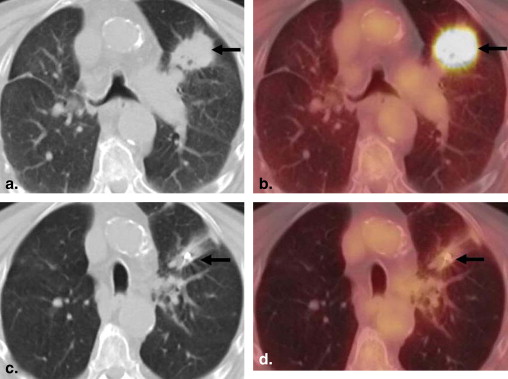
PET/CT Imaging
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
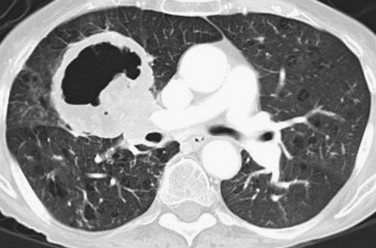
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Multiparametric Approach for Tumor Response Assessment
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Jemal A., Siegel R., Ward E., et. al.: Cancer statistics, 2009. CA Cancer J Clin 2009; 59: pp. 225-249.
2. American Cancer Society. Cancer facts and figures, 2009. Available at: http://ww3.cancer.org/downloads/STT/500809web.pdf . Accessed December 22, 2010.
3. Non-Small Cell Lung Cancer Collaborative Group: Chemotherapy in non-small cell lung cancer: a meta-analysis using updated data on individual patients from 52 randomised clinical trials. BMJ 1995; 311: pp. 899-909.
4. Breathnach O.S., Freidlin B., Conley B., et. al.: Twenty-two years of phase III trials for patients with advanced non-small-cell lung cancer: sobering results. J Clin Oncol 2001; 19: pp. 1734-1742.
5. Lynch T.J., Bell D.W., Sordella R., et. al.: Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350: pp. 2129-2139.
6. Paez J.G., Janne P.A., Lee J.C., et. al.: EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: pp. 1497-1500.
7. Pao W., Miller V., Zakowski M., et. al.: EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 2004; 101: pp. 13306-13311.
8. Gazdar A.: Personalized medicine and inhibition of EGFR signaling in lung cancer. N Engl J Med 2009; 361: pp. 1018-1020.
9. Herbst R.S., Heymach J.V., Lippman S.M.: Lung cancer. N Engl J Med 2008; 359: pp. 1367-1380.
10. Sun S., Schiller J.H., Gazdar A.F.: Lung cancer in never smokers—a different disease. Nat Rev Cancer 2007; 7: pp. 778-790.
11. Sato M., Shames D.S., Gazdar A.F., et. al.: A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol 2007; 2: pp. 327-343.
12. Brabender J., Danenberg K.D., Metzger R., et. al.: Epidermal growth factor receptor and HER2-neu mRNA expression in non-small cell lung cancer is correlated with survival. Clin Cancer Res 2001; 7: pp. 1850-1855.
13. Rusch V., Klimstra D., Venkatraman E., et. al.: Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin Cancer Res 1997; 3: pp. 515-522.
14. Fukuoka M., Yano S., Giaccone G., et. al.: Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (the IDEAL 1 trial). [published erratum appears in J Clin Oncol 2004; 22:4811] J Clin Oncol 2003; 21: pp. 2237-2246.
15. Kris M.G., Natale R.B., Herbst R.S., et. al.: Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA 2003; 290: pp. 2149-2158.
16. Pérez-Soler R., Chachoua A., Hammond L.A., et. al.: Determinants of tumor response and survival with erlotinib in patients with non–small-cell lung cancer. J Clin Oncol 2004; 22: pp. 3238-3247.
17. van Zandwijk N., Mathy A., Boerrigter L., et. al.: EGFR and KRAS mutations as criteria for treatment with tyrosine kinase inhibitors: retro- and prospective observations in non-small-cell lung cancer. Ann Oncol 2007; 18: pp. 99-103.
18. Sequist L.V., Martins R.G., Spigel D., et. al.: First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol 2008; 26: pp. 2442-2449.
19. Jackman D.M., Yeap B.Y., Sequist L.V., et. al.: Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res 2006; 12: pp. 3908-3914.
20. Inoue A., Suzuki T., Fukuhara T., et. al.: Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol 2006; 24: pp. 3340-3346.
21. Asahina H., Yamazaki K., Kinoshita I., et. al.: A phase II trial of gefitinib as firstline therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer 2006; 95: pp. 998-1004.
22. Sequist L.V., Settleman J.E., Ackman J.B., et. al.: Case records of the Massachusetts General Hospital. Case 23-2008. A 26-year-old man with back pain and a mass in the lung. N Engl J Med 2008; 359: pp. 405-414.
23. Mok T.S., Wu Y.L., Thongprasert S., et. al.: Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: pp. 947-957.
24. Jackman D.M., Miller V.A., Cioffredi L.A., et. al.: Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res 2009; 15: pp. 5267-5273.
25. Shepherd F.A., Rodrigues Pereira J., Ciuleanu T., et. al.: Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005; 353: pp. 123-132.
26. Rodenhuis S., Slebos R.J.: Clinical significance of ras oncogene activation in human lung cancer. Cancer Res 1992; 52: pp. 2665s-2669s.
27. Riely G.J., Marks J., Pao W.: KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc 2009; 6: pp. 201-205.
28. Jänne P.A., Engelman J.A., Johnson B.E.: Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol 2005; 23: pp. 3227-3234.
29. Jackman D.M.: EGFR biomarkers: predicting sensitivity to therapy. Mol Oncol Rep 2006; 1: pp. 31-36.
30. Thatcher N., Chang A., Parikh P., et. al.: Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer). Lancet 2005; 366: pp. 1527-1537.
31. Shigematsu H., Lin L., Takahashi T., et. al.: Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005; 97: pp. 339-346.
32. Tsao M.S., Sakurada A., Cutz J.C., et. al.: Erlotinib in lung cancer—molecular and clinical predictors of outcome. [published erratum appears in N Engl J Med 2006; 355:1746] N Engl J Med 2005; 353: pp. 133-144.
33. Hirsch F.R., Varella-Garcia M., McCoy J., et. al.: Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group study. J Clin Oncol 2005; 23: pp. 6838-6845.
34. Cappuzzo F., Hirsch F.R., Rossi E., et. al.: Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 2005; 97: pp. 643-655.
35. Solomon B., Gregorc V., Taguchi F., et. al.: Prediction of clinical outcome in non-small-cell lung cancer (NSCLC) patients treated with gefitinib using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-ToF) of serum. J Clin Oncol 2006; 24: pp. 7004.
36. Pao W., Wang T.Y., Riely G.J., et. al.: KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2005; 2: pp. e17.
37. Eberhard D.A., Johnson B.E., Amler L.C., et. al.: Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005; 23: pp. 5900-5909.
38. Han S.W., Kim T.Y., Jeon Y.K., et. al.: Optimization of patient selection for gefitinib in non-small cell lung cancer by combined analysis of epidermal growth factor receptor mutation, K-ras mutation, and Akt phosphorylation. Clin Cancer Res 2006; 12: pp. 2538-2544.
39. Kobayashi S., Boggon T.J., Dayaram T., et. al.: EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005; 24: pp. 786-792.
40. Pao W., Miller V.A., Politi K.A., et. al.: Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005; 2: pp. e73.
41. Bell D.W., Gore I., Okimoto R.A., et. al.: Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005; 37: pp. 1315-1316.
42. Engelman J.A., Zejnullahu K., Mitsudomi T., et. al.: MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316: pp. 1039-1043.
43. Bean J., Brennan C., Shih J.Y., et. al.: MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007; 104: pp. 20932-20937.
44. Jackman D., Pao W., Riely G.J., et. al.: Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol 2010; 28: pp. 357-360.
45. McDermott U., Settleman J.: Personalized cancer therapy with selective kinase inhibitors: an emerging paradigm in medical oncology. J Clin Oncol 2009; 27: pp. 5650-5659.
46. Rosell R., Moran T., Queralt C., et. al.: Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361: pp. 958-967.
47. Ding L., Johnson B.E., Meyerson M., et. al.: Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: pp. 1069-1075.
48. Bass A.J., Watanabe H., Mermel C.H., et. al.: SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009; 41: pp. 1238-1242.
49. Ohtsuka K., Ohnishi H., Fujiwara M., et. al.: Abnormalities of epidermal growth factor receptor in lung squamous-cell carcinomas, adenosquamous carcinomas, and large-cell carcinomas: tyrosine kinase domain mutations are not rare in tumors with an adenocarcinoma component. Cancer 2007; 109: pp. 741-750.
50. Travis W., Nicholson S., Hirsch F.R., et. al.: Tumours of the lung. Tumours of the lung, pleura, thymus and heart. World Health Organization Classification of Tumours. Pathology and Genetics.2004.IARCLyon, France 31–34
51. Kitamura H., Yazawa T., Okudela K., et. al.: Molecular and genetic pathogenesis of lung cancer: differences between small-cell and non-small-cell carcinomas. Open Pathol J 2008; 2: pp. 106-114.
52. Kim Y.H., Girard L., Giacomini C.P., et. al.: Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene 2006; 25: pp. 130-138.
53. Zakowski M.F., Hussain S., Pao W., et. al.: Morphologic features of adenocarcinoma of the lung predictive of response to the epidermal growth factor receptor kinase inhibitors erlotinib and gefitinib. Arch Pathol Lab Med 2009; 133: pp. 470-477.
54. Weir B.A., Johnson B.E., Meyerson M., et. al.: Characterizing the cancer genome in lung adenocarcinoma. Nature 2007; 450: pp. 893-898.
55. World Health Organization. WHO Handbook for Reporting Results of Cancer Treatment. Available at: http://whqlibdoc.who.int/publications/9241700483.pdf . Accessed December 22, 2010.
56. Therasse P., Arbuck S.G., Eisenhauer E.A., et. al.: New guidelines to evaluate the response to treatment in solid tumors: European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000; 92: pp. 205-216.
57. Eisenhauer E.A., Therasse P., Bogaerts J., et. al.: New response evaluation criteria in solid tumors: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: pp. 228-247.
58. Suzuki C., Jacobsson H., Hatschek T., et. al.: Radiologic measurements of tumor response to treatment: practical approaches and limitations. Radiographics 2008; 28: pp. 329-344.
59. Gavrielides M.A., Kinnard L.M., Myers K.J., et. al.: Noncalcified lung nodules: volumetric assessment with thoracic CT. Radiology 2009; 251: pp. 26-37.
60. Zhao B., Schwartz L.H., Moskowitz C.S., et. al.: Pulmonary metastases: effect of CT section thickness on measurement—initial experience. Radiology 2005; 234: pp. 934-939.
61. Zhao B., Schwartz L.H., Moskowitz C.S., et. al.: Lung cancer: computerized quantification of tumor response—initial results. Radiology 2006; 241: pp. 892-898.
62. Zhao B., James L.P., Moskowitz C.S., et. al.: Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology 2009; 252: pp. 263-272.
63. Yankelevitz D.F., Reeves A.P., Kostis W.J., et. al.: Small pulmonary nodules: volumetrically determined growth rates based on CT evaluation. Radiology 2000; 217: pp. 251-256.
64. Kostis W.J., Yankelevitz D.F., Reeves A.P., et. al.: Small pulmonary nodules: reproducibility of three-dimensional volumetric measurement and estimation of time to follow-up CT. Radiology 2004; 231: pp. 446-452.
65. van Klaveren R.J., Oudkerk M., Prokop M., et. al.: Management of lung nodules detected by volume CT scanning. N Engl J Med 2009; 361: pp. 2221-2229.
66. Marom E.M., Martinez C.H., Truong M.T., et. al.: Tumor cavitation during therapy with antiangiogenesis agents in patients with lung cancer. J Thorac Oncol 2008; 3: pp. 351-357.
67. Choyke P.L., Dwyer A.J., Knopp M.V.: Functional tumor imaging with dynamic contrast-enhanced magnetic resonance imaging. J Magn Reson Imaging 2003; 17: pp. 509-520.
68. Hatabu H., Gaa J., Kim D., et. al.: Pulmonary perfusion: qualitative assessment with dynamic contrast-enhanced MRI using ultra-short TE and inversion recovery turbo FLASH. Magn Reson Med 1996; 36: pp. 503-508.
69. Hatabu H., Tadamura E., Levin D.L., et. al.: Quantitative assessment of pulmonary perfusion with dynamic contrast-enhanced MRI. Magn Reson Med 1999; 42: pp. 1033-1038.
70. Ohno Y., Hatabu H., Takenaka D., Adachi S., Kono M., Sugimura K.: Solitary pulmonary nodules: potential role of dynamic MR imaging in management initial experience. Radiology 2002; 224: pp. 503-511.
71. Schaefer J.F., Vollmar J., Schick F., et. al.: Solitary pulmonary nodules: dynamic contrast-enhanced MR imaging—perfusion differences in malignant and benign lesions. Radiology 2004; 232: pp. 544-553.
72. Donmez F.Y., Yekeler E., Saeidi V., Tunaci A., Tunaci M., Acunas G.: Dynamic contrast enhancement patterns of solitary pulmonary nodules on 3D gradient-recalled echo MRI. AJR Am J Roentgenol 2007; 189: pp. 1380-1386.
73. Zou Y., Zhang M., Wang Q., Shang D., Wang L., Yu G.: Quantitative investigation of solitary pulmonary nodules: dynamic contrast-enhanced MRI and histopathologic analysis. AJR Am J Roentgenol 2008; 191: pp. 252-259.
74. Fujimoto K., Abe T., Müller N.L., et. al.: Small peripheral pulmonary carcinomas evaluated with dynamic MR imaging: correlation with tumor vascularity and prognosis. Radiology 2003; 227: pp. 786-793.
75. Ohno Y., Nogami M., Higashino T., et. al.: Prognostic value of dynamic MR imaging for non-small-cell lung cancer patients after chemoradiotherapy. J Magn Reson Imaging 2005; 21: pp. 775-783.
76. Warburg O.: On the origin of cancer cells. Science 1956; 123: pp. 309-314.
77. Flier J.S., Mueckler M.M., Usher P., et. al.: Elevated levels of glucose transport and transport messenger RNA are induced by ras or src oncogenes. Science 1987; 235: pp. 1492-1495.
78. Weber W.A., Figlin R.: Monitoring cancer treatment with PET/CT: does it make a difference?. J Nucl Med 2007; 48: pp. 36S-44S.
79. Thie J.A.: Understanding the standardized uptake value, its methods, and implications for usage. J Nucl Med 2004; 45: pp. 1431-1434.
80. Patz E.F., Lowe V.J., Hoffman J.M., et. al.: Focal pulmonary abnormalities: evaluation with F-18 fluorodeoxyglucose PET scanning. Radiology 1993; 188: pp. 487-490.
81. Lowe V.J., Fletcher J.W., Gobar L., et. al.: Prospective investigation of positron emission tomography in lung nodules. J Clin Oncol 1998; 16: pp. 1075-1084.
82. Young H., Baum R., Cremerius U., et. al., European Organization for Research and Treatment of Cancer (EORTC) PET Study Group: Measurement of clinical and subclinical tumour response using [18F]-fluorodeoxyglucose and positron emission tomography: review and 1999 EORTC recommendations. Eur J Cancer 1999; 35: pp. 1773-1782.
83. de Geus-Oei L.F., van der Heijden H.F., Corstens F.H., et. al.: Predictive and prognostic value of FDG-PET in nonsmall-cell lung cancer: a systematic review. Cancer 2007; 110: pp. 1654-1664.
84. Nahmias C., Hanna W.T., Wahl L.M., et. al.: Time course of early response to chemotherapy in non-small cell lung cancer patients with 18F-FDG PET/CT. J Nucl Med 2007; 48: pp. 744-751.
85. Wahl R.L., Jacene H., Kasamon Y., et. al.: From RECIST to PERCIST; evolving considerations for PET response criteria in solid tumors. J Nucl Med 2009; 50: pp. 122S-150S.
86. Shankar L.K., Sullivan D.C.: Functional imaging in lung cancer. J Clin Oncol 2005; 23: pp. 3203-3211.
87. Zhao B., Schwartz L.H., Larson S.M.: Imaging surrogates of tumor response to therapy: anatomic and functional biomarkers. J Nucl Med 2009; 50: pp. 239-249.
88. Buck A.K., Schirrmeister H., Hetzel M., et. al.: 3-deoxy-3-[(18)F]fluorothymidine-positron emission tomography for noninvasive assessment of proliferation in pulmonary nodules. Cancer Res 2002; 62: pp. 3331-3334.
89. Padhani A.R., Miles K.A.: Multiparametric imaging of tumor response to therapy. Radiology 2010; 256: pp. 348-364.
90. Groves A.M., Wishart G.C., Shastry M., et. al.: Metabolic-flow relationships in primary breast cancer: feasibility of combined PET/dynamic contrast-enhanced CT. Eur J Nucl Med Mol Imaging 2009; 36: pp. 416-421.
91. Jeong Y.J., Lee K.S., Jeong S.Y., et. al.: Solitary pulmonary nodule: characterization with combined wash-in and washout features at dynamic multi-detector row CT. Radiology 2005; 237: pp. 675-683.
92. Ng Q.S., Goh V., Milner J., et. al.: Quantitative helical dynamic contrast enhanced computed tomography assessment of the spatial variation in whole tumour blood volume with radiotherapy in lung cancer. Lung Cancer 2010; 69: pp. 71-76.
93. Lazanyi K.S., Abramyuk A., Wolf G., et. al.: Usefulness of dynamic contrast enhanced computed tomography in patients with non-small-cell lung cancer scheduled for radiation therapy. Lung Cancer 2010; 70: pp. 280-285.
94. Lind J.S., Meijerink M.R., Dingemans A.M., et. al.: Dynamic contrast-enhanced CT in patients treated with sorafenib and erlotinib for non-small cell lung cancer: a new method of monitoring treatment?. Eur Radiol 2010; 20: pp. 2890-2898.
95. Van Oosterom A.T., Dumez H., Desai J., et. al.: Combination signal transduction inhibition: a phase I/II trial of the oral mTOR inhibitor everolimus (E, RAD001) and imatinib mesylate (IM) in patients (pts) with gastrointestinal stromal tumor (GIST) refractory to IM. J Clin Oncol 2004; 22: pp. 3002.
96. Riely G.J., Kris M.G., Zhao B., et. al.: Prospective assessment of discontinuation and reinitiation of erlotinib or gefitinib in patients with acquired resistance to erlotinib or gefitinib followed by the addition of everolimus. Clin Cancer Res 2007; 13: pp. 5150-5155.