Rationale and Objectives
This article presents an initiative for the translation of advances in neuroimage analysis techniques to clinical research scientists. Our objective is to bridge the gap between scientific advances made by the biomedical imaging community and their widespread use in the clinical research community. Through national collaborative effort supported by the National Institutes of Health Roadmap, the integration of the most sophisticated algorithms into usable working open-source systems enables clinical researchers to have access to a broad spectrum of cutting edge analysis techniques. A critical step to maximize the long-term positive impact of this collaborative effort is to translate these techniques into new skills of clinical researchers. To address this challenge, we developed a methodology based on three criteria: a multidisciplinary approach, a balance between theory and common practice, and an immersive collaborative environment. The article illustrates our initiative through the exemplar case of diffusion tensor imaging tractography, and reports on our experience over the past two years of designing and delivering training workshops to more than 300 clinicians and scientists using the developed methodology.
The rapid development and application of neuroimaging over the past 20 years has led to a better understanding of brain function in health and disease. New techniques such as magnetic resonance imaging and including diffusion tensor magnetic resonance imaging (DTI) provide powerful capabilities for noninvasive studies of the brain ( ). Image analysis techniques also play a critical role in the extraction of meaningful information that is relevant to biomarkers for diagnosis, prognosis, and treatment response. Moreover, the emergence of increasingly sophisticated mathematic models, image processing, and visualization tools enables the comprehensive mapping of brain structure and function.
Publications in peer-reviewed journals and scientific conferences provide dissemination of new scientific principles, and deployment of the image processing algorithms, including support that enables reusability, is hampered by many factors. These include the lack of calibration and validation of the techniques required to achieve meaningful reproducibility of results. Another factor is the many sources of variability in neuroimage data. The benefits of the developed algorithms can be limited by a lack of support for finish engineering, such as providing for multiplatform support, upgrades, and training materials. The major part of the development of new image analysis tools within the medical imaging community occurs at a local scale, based on focused efforts of individual research groups. As a result, valuable noncommercial software is blocked from reaching the broader scientific and clinical research communities.
Many of these obstacles have been identified by multiple groups as part of a joint effort toward the identification and characterization of the opportunities for scientific research and engineering development in biomedical imaging ( ). To solve these challenges, the National Institutes of Health (NIH) Roadmap initiative has laid out a vision for a more efficient and productive system of medical research. Essential elements in the NIH Roadmap and the research pipeline are the translational steps, and global consortia such as the National Alliance for Medical Image Computing (NA-MIC) and the Center for Computational Biology (CCB), two of the seven National Centers for Biomedical Computing, focus their efforts on the conversion of scientific advances from the biomedical imaging community into working open-source systems, to improve the availability and deployment of these tools on a national scale ( ). The NA-MIC consortium involves clinical researchers in the definition of the tools that will address their hypotheses, computer scientists in the design and implementation of the corresponding algorithms, and engineers for ensuring robustness and usability of the open-source software toolset made available to the broader community. This movement in general, and the NA-MIC consortium in particular, builds on the open-source effort such as the National Library of Medicine’s Insight Toolkit ( ), the Visualization Toolkit ( ), and the CMake build environment ( ). These efforts include the integration of the best available practices in software development, including architecture design, software engineering process, and automated quality assurance. Cutting-edge biomedical computing algorithms thus become accessible through end-user applications compatible with major computer platforms.
Still, such technology and knowledge-sharing systems face the challenge of translating scientific advances made by engineers and computer scientists to a broader community of clinical research scientists. It is not enough to make a new technology accessible, its deployment must be converted into new skills for these scientists and clinicians, and the most sophisticated mathematical algorithms will achieve little impact on biomedical discoveries if they do not reach the clinical researchers for whom they were developed.
Potential solutions must therefore be implemented to bridge this gap and ensure that clinical researchers are able to use new tools in their investigations. In this article, we propose a perspective for translating advances in neuroimage analysis to clinical researchers.
Requirements for Transferring Advances in Neuroimage Analysis
Get Radiology Tree app to read full this article<
Multidisciplinary
Get Radiology Tree app to read full this article<
Theory into practice
Get Radiology Tree app to read full this article<
Immersion in a collaborative environment
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
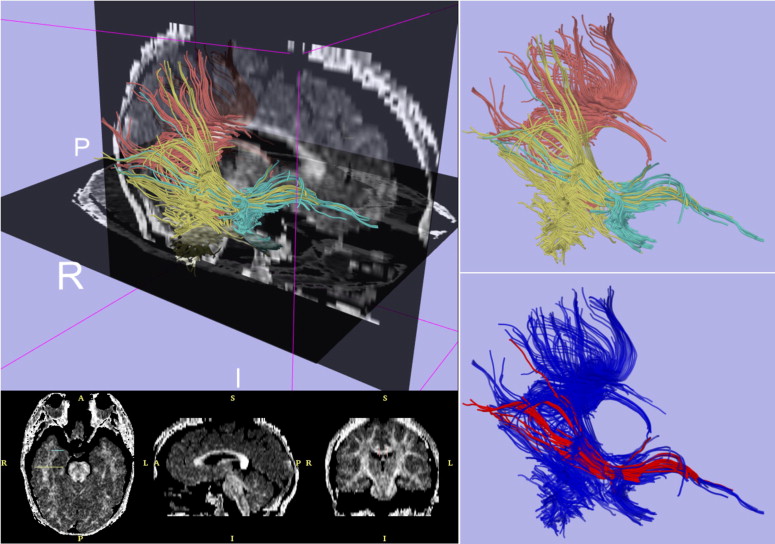
Exemplar Case: Diffusion Tensor Imaging Tractography
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusion and perspectives
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Acknowledgments
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Basser P.J., Mattiello J., LeBihan D.: MR diffusion tensor spectroscopy and imaging. Biophys J 1994; 66: pp. 259-267.
2. Basser P.J., Pajevic S., Pierpaoli C., et. al.: In vivo fiber tractography using DT-MRI data. Magn Reson Med 2000; 44: pp. 625-632.
3. Carson P.L., Giger M., Welch M.J., et. al.: Biomedical Imaging Research Opportunities Workshop: report and recommendations. Radiology 2003; 229: pp. 328-339.
4. Hendee W.R.: Biomedical Imaging Research Opportunities Workshop II: a summary of findings and recommendations. Med Phys 2005; 32: pp. 2484-2486.
5. Hendee W.R.: Biomedical Imaging Research Opportunities Workshop III: summary of findings and recommendations. Radiology 2006; 238: pp. 402-404.
6. Hendee W.R.: Special report: biomedical imaging research opportunities workshop IV—a summary of findings and recommendations. Radiology 2007; 242: pp. 338-341.
7. National Alliance for Medical Image Computing. Available online at http://www.na-mic.org
8. Center for Computational Biology at UCLA. Available online at http://www.loni.ucla.edu/CCB/
9. National Institutes of Health. NIH Roadmap National Centers for Biomedical Computing. Available online at http://www.bisti.nih.gov/ncbc/
10. Ibanez L., Schroeder W., Ng L., et. al.: The ITK software guide.2003.Kitware IncClifton Park, NY
11. Schroeder W., Martin K., Lorensen W.: The visualization toolkit.ed 3.2004.Kitware IncClifton Park, NY
12. Martin K., Hoffman B.: Mastering Cmake.ed 2.2006.Kitware IncClifton Park, NY
13. Kubicki M., Park H., Westin C.F., et. al.: DTI and MTR abnormalities in schizophrenia: analysis of white matter integrity. Neuroimage 2005; 26: pp. 1109-1118.
14. Burns J., Job D., Bastin M.E., et. al.: Structural disconnectivity in schizophrenia: a diffusion tensor magnetic resonance imaging study. Br J Psychiatry 2003; 182: pp. 439-443.
15. 3D Slicer. Available online at http://www.slicer.org
16. NIH Roadmap. National Centers for Biomedical Computing. 2006 All Hands Meeting. Available online at http://www.bisti.nih.gov/ahm2006/index.htm
17. NA-MIC Training Compendium. Available online at http://www.na-mic.org/Wiki/index.php/Slicer:Workshops:User_Training_101