Rationale and Objectives
To investigate the effect of a computer-aided diagnosis (CADx) system on radiologists’ performance in discriminating malignant and benign masses on mammograms and three-dimensional (3D) ultrasound (US) images.
Materials and Methods
Our dataset contained mammograms and 3D US volumes from 67 women (median age, 51; range: 27–86) with 67 biopsy-proven breast masses (32 benign and 35 malignant). A CADx system was designed to automatically delineate the mass boundaries on mammograms and the US volumes, extract features, and merge the extracted features into a multi-modality malignancy score. Ten experienced readers (subspecialty academic breast imaging radiologists) first viewed the mammograms alone, and provided likelihood of malignancy (LM) ratings and Breast Imaging and Reporting System assessments. Subsequently, the reader viewed the US images with the mammograms, and provided LM and action category ratings. Finally, the CADx score was shown and the reader had the opportunity to revise the ratings. The LM ratings were analyzed using receiver-operating characteristic (ROC) methodology, and the action category ratings were used to determine the sensitivity and specificity of cancer diagnosis.
Results
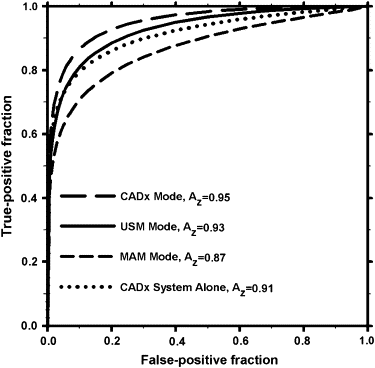
Without CADx, readers’ average area under the ROC curve, A z , was 0.93 (range, 0.86–0.96) for combined assessment of the mass on both the US volume and mammograms. With CADx, their average A z increased to 0.95 (range, 0.91–0.98), which was borderline significant ( P = .05). The average sensitivity of the readers increased from 98% to 99% with CADx, while the average specificity increased from 27% to 29%. The change in sensitivity with CADx did not achieve statistical significance for the individual radiologists, and the change in specificity was statistically significant for one of the radiologists.
Conclusions
A well-trained CADx system that combines features extracted from mammograms and US images may have the potential to improve radiologists’ performance in distinguishing malignant from benign breast masses and making decisions about biopsies.
Breast cancer is the second leading cause of cancer death and the most prevalent noncutaneous cancer among American women . Because early detection of breast cancer may improve the chance of survival, Breast Imaging Reporting and Data System (BI-RADS) categories 4 and 5 findings are typically referred for biopsy. Although the optimal positive biopsy rate for abnormalities detected by mammography is still being debated, positive breast biopsy rates of 25%–40% have been recommended as appropriate . Studies indicate that academic and community practices perform close to the lower end of this recommendation .
Increasing the positive predictive value of biopsy would reduce the cost of health care, spare patients the anxiety and discomfort of biopsy, and avoid the possible scarring of the breast tissue, which might complicate future exams. However, this increased positive predictive value should not come at the cost of missed cancers, but rather as a result of an overall improvement in the accuracy of breast cancer detection and characterization. Computer-aided diagnosis (CADx) is one of the techniques that strive to improve radiologists’ differentiation between malignant and benign lesions, thus improving the positive biopsy rate without missing malignancies.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and methods
Dataset
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
CADx System
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Observer Performance Study
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Data and Statistical Analysis
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Results
Get Radiology Tree app to read full this article<
Table 1
The Area A z under ROC Curve, and the Partial Area Index above a Sensitivity of 0.9, A z (0.9) , for Characterization of the Masses in our Dataset in MAM, USM, and CADx Reading Modes
Area A z under ROC Curve The Partial Area Index above a Sensitivity of 0.9, A z (0.9) Radiologist Years of Experience MAM Mode USM Mode CADx Mode MAM Mode USM Mode CADx Mode 1 3 0.88 ± 0.04 0.96 ± 0.02 0.98 ± 0.01 0.23 ± 0.15 0.43 ± 0.17 0.52 ± 0.15 2 10 0.83 ± 0.05 0.86 ± 0.04 0.91 ± 0.04 0.30 ± 0.14 0.42 ± 0.13 0.63 ± 0.21 3 26 0.87 ± 0.04 0.92 ± 0.04 0.96 ± 0.02 0.54 ± 0.16 0.77 ± 0.19 0.72 ± 0.13 4 20 0.84 ± 0.05 0.92 ± 0.03 0.95 ± 0.03 0.38 ± 0.12 0.66 ± 0.16 0.83 ± 0.11 5 6 0.93 ± 0.03 0.96 ± 0.02 0.96 ± 0.02 0.04 ± 0.18 0.49 ± 0.10 0.60 ± 0.18 6 5 0.87 ± 0.04 0.95 ± 0.03 0.98 ± 0.02 0.41 ± 0.15 0.64 ± 0.19 0.55 ± 0.20 7 17 0.90 ± 0.04 0.95 ± 0.03 0.95 ± 0.03 0.35 ± 0.16 0.40 ± 0.17 0.53 ± 0.14 8 14 0.84 ± 0.07 0.93 ± 0.03 0.95 ± 0.03 0.36 ± 0.15 0.62 ± 0.19 0.81 ± 0.13 9 12 0.80 ± 0.06 0.87 ± 0.05 0.91 ± 0.03 0.07 ± 0.06 0.50 ± 0.15 0.71 ± 0.10 10 10 0.85 ± 0.05 0.90 ± 0.04 0.93 ± 0.03 0.02 ± 0.14 0.27 ± 0.17 0.55 ± 0.14 Average 12 0.87 0.93 0.95 0.27 0.52 0.65P value .03 .05 .001 .008
ROC: receiver-operating characteristic; MAM: mammography; USM: ultrasound and mammography; CADx: computer-aided diagnosis; MRMC: multireader multicase.
The P values of the difference in average A z value observed in USM mode compared to MAM mode, and in CADx mode compared to USM mode were estimated using MRMC ROC analysis. The corresponding P values for A z (0.9) were estimated using two-tailed paired t -test.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 2
Numbers of Benign and Malignant Masses for which the 10 Radiologists’ LM Rating Changed at Step 2 (MAM→USM) and Step 3 (USM→CADx) in the Observer Study
MAM→USM USM→CADx LM changes for malignant masses Beneficial 190 (54) 97 (28) Detrimental 20 (6) 11 (3) Total 210 (60) 108 (31) LM changes for benign masses Beneficial 104 (33) 74 (23) Detrimental 110 (34) 56 (18) Total 214 (67) 130 (41) Total changes Beneficial 294 (44) 171 (26) Detrimental 130 (19) 67 (10) Total 424 (63) 238 (36)
LM: likelihood of malignancy; MAM: mammography; USM: ultrasound and mammography; CADx: computer-aided diagnosis.
Numbers in parentheses are percentages. The total number of readings for malignant masses was 350 (35 masses × 10 radiologists) and the total number of readings for benign masses was 320 (32 masses × 10 radiologists). A beneficial change for a malignant mass is an increase in the LM rating, and that for a benign mass is a decrease in the LM rating.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 3
The Sensitivity and Specificity for Each Radiologist
MAM Mode USM Mode CADx Mode Radiologist Sensitivity Specificity Sensitivity Specificity Sensitivity Specificity 1 0.97 0.34 1.00 0.16 1.00 0.16 2 0.83 0.66 0.89 0.66 0.94 0.63 3 1.00 0.13 1.00 0.13 1.00 0.19 4 0.91 0.41 0.97 0.34 1.00 0.34 5 1.00 0.16 1.00 0.16 1.00 0.28 6 0.91 0.44 0.97 0.44 1.00 0.34 7 1.00 0.13 1.00 0.09 0.97 0.28 8 0.77 0.84 0.94 0.53 0.97 0.50 9 1.00 0.00 1.00 0.09 1.00 0.09 10 1.00 0.25 1.00 0.13 1.00 0.09 Average 0.94 0.33 0.98 0.27 0.99 0.29P value 0.06 0.13 0.17 0.50
MAM: mammography; USM: ultrasound and mammography; CADx: computer-aided diagnosis.
The significances of the difference in the averages between USM mode vs. MAM mode, and CADx mode vs, USM mode was estimated using Student’s paired t -test.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 4
Changes in Biopsy Decisions for Each Radiologist in Step 2 (MAM→USM,) and Step 3 (USM→CADx) in the Observer Study
Change in Biopsy Decision, Step 2 (MAM→USM) Change in Biopsy Decision, Step 3 (USM→CADx) Malignant Masses Benign Masses Malignant Masses Benign Masses Radiologist Beneficial Detrimental Beneficial Detrimental Beneficial Detrimental Beneficial Detrimental 1 1 0 1 7 0 0 0 0 2 4 2 4 4 2 0 1 2 3 0 0 2 2 0 0 2 0 4 3 1 4 6 1 0 1 1 5 0 0 3 3 0 0 5 1 6 3 1 6 6 1 0 1 4 7 0 0 2 3 0 1 6 † 0 8 6 ∗ 0 1 † 11 1 0 3 4 9 0 0 3 0 0 0 1 1 10 0 0 2 6 0 0 0 1 Total 17 4 28 48 5 1 20 14
LM: likelihood of malignancy; MAM: mammography; USM: ultrasound and mammography; CADx: computer-aided diagnosis.
A beneficial change for a malignant mass is a decision to recommend biopsy for the mass that was not recommended for biopsy in the previous step. A beneficial change for a benign mass is a decision not to recommend biopsy for the mass that was recommended for biopsy in the previous step.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 5
Changes in Biopsy Decisions for Each Radiologist in Step 3 (USM→CADx) if the Decision Threshold for Biopsy Recommendation were Chosen to be 10% in CADx Mode, and Maintained at 2% in USM Mode, with the Sensitivity Fixed at 98% for Both Modes
Change in Biopsy Decision for Malignant Mass Change in Biopsy Decision for Benign Mass Radiologist Beneficial Detrimental Beneficial Detrimental 1 ∗ 0 0 7 0 2 2 1 1 1 3 0 0 4 0 4 1 0 2 1 5 † 0 0 9 1 6 1 0 1 3 7 † 0 1 12 0 8 1 1 3 3 9 0 0 4 1 10 0 0 4 1 Total 5 3 47 11
LM: likelihood of malignancy; MAM: mammography; USM: ultrasound and mammography; CADx: computer-aided diagnosis.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Jemal A., Siegel J., Ward E., et. al.: Cancer statistics, 2007. CA Cancer J Clin 2007; 57: pp. 43-66.
2. Bassett L.W., Hendrick R.E., Bassford T.L., et. al.: Quality determinants of mammography. Clinical Practice Guideline No. 13. AHCPR Publication No. 95–0632.1994.Agency for Health Care Policy and Research, Public Health Service, US Department of Health and Human ServicesRockville, MD
3. Brown M.L., Houn F., Sickles E.A., et. al.: Screening mammography in community practice: positive predictive value of abnormal findings and yield of follow-up diagnostic procedures. AJR Am J Roentgenol 1995; 165: pp. 1373-1377.
4. Gur D., Wallace L.P., Klym A.H., et. al.: Trends in recall, biopsy, and positive biopsy rates for screening mammography in an academic practice. Radiology 2005; 235: pp. 396-401.
5. Huo Z.M., Giger M.L., Vyborny C.J., et. al.: Computerized classification of benign and malignant masses on digitized mammograms: a study of robustness. Acad Radiol 2000; 7: pp. 1077-1084.
6. Mudigonda N.R., Rangayyan R.M., Desautels J.E.L.: Gradient and texture analysis for the classification of mammographic masses. IEEE Trans Med Imaging 2000; 19: pp. 1032-1043.
7. Leichter I., Fields S., Nirel R., et. al.: Improved mammographic interpretation of masses using computer-aided diagnosis. Eur Radiol 2000; 10: pp. 377-383.
8. Bilska-Wolak A.O., Floyd C.E., Nolte L.W., et. al.: Application of likelihood ratio to classification of mammographic masses; performance comparison to case-based reasoning. Med Phys 2003; 30: pp. 949-958.
9. Varela C., Timp S., Karssemeijer N.: Use of border information in the classification of mammographic masses. Phys Medic Biol 2006; 51: pp. 425-441.
10. Cheng H.D., Shi X.J., Min R., et. al.: Approaches for automated detection and classification of masses in mammograms. Pattern Recognit 2006; 39: pp. 646-668.
11. Delogu P., Fantacci M.E., Kasae P., et. al.: Characterization of mammographic masses using a gradient-based segmentation algorithm and a neural classifier. Comp Biol Med 2007; 37: pp. 1479-1491.
12. Shi J., Sahiner B., Chan H.P., et. al.: Characterization of mammographic masses based on level set segmentation with new image features and patient information. Med Phys 2008; 35: pp. 280-290.
13. Chen D.R., Chang R.F., Huang Y.L.: Computer-aided diagnosis applied to US of solid breast nodules by using neural networks. Radiology 1999; 213: pp. 407-412.
14. Dumane V.A., Shankar P.M., Piccoli C.W., et. al.: Computer aided classification of masses in ultrasonic mammography. Med Phys 2002; 29: pp. 1968-1973.
15. Chen C.M., Chou Y.H., Han K.C., et. al.: Breast lesions on sonograms: computer-aided diagnosis with nearly setting-independent features and artificial neural networks. Radiology 2003; 226: pp. 504-514.
16. Horsch K., Giger M.L., Vyborny C.J., et. al.: Performance of computer-aided diagnosis in the interpretation of lesions on breast sonography. Acad Radiol 2004; 11: pp. 272-280.
17. Joo S., Yang Y.S., Moon W.K., et. al.: Computer-aided diagnosis of solid breast nodules: use of an artificial neural network based on multiple sonographic features. IEEE Trans Med Imaging 2004; 23: pp. 1292-1300.
18. Sahiner B., Chan H.P., Roubidoux M.A., et. al.: Computerized characterization of breast masses on 3-D ultrasound volumes. Med Phys 2004; 31: pp. 744-754.
19. Sehgal C.M., Cary T.W., Kangas S.A., et. al.: Computer-based margin analysis of breast sonography for differentiating malignant and benign masses. J Ultrasound Med 2004; 23: pp. 1201-1209.
20. Shen W.C., Chang R.F., Moon W.K., et. al.: Breast ultrasound computer-aided diagnosis using BI-RADS features. Acad Radiol 2007; 14: pp. 928-939.
21. Sahiner B., Chan H.P., Hadjiiski L.M., et. al.: Fusion of mammographic and sonographic computer-extracted features for improved characterization of breast masses. Proc 7th Int Workshop Digital Mammogr 2004; pp. 199-202.
22. Drukker K., Horsch K., Giger M.L.: Multimodality computerized diagnosis of breast lesions using mammography and sonography. Acad Radiol 2005; 12: pp. 970-979.
23. Jesneck J.L., Lo J.Y., Baker J.A.: Breast mass lesions: computer-aided diagnosis models with mammographic and sonographic descriptors. Radiology 2007; 244: pp. 390-398.
24. Chan H.P., Sahiner B., Helvie M.A., et. al.: Improvement of radiologists’ characterization of mammographic masses by computer-aided diagnosis: an ROC study. Radiology 1999; 212: pp. 817-827.
25. Markey M.K., Lo J.Y., Floyd C.E.: Differences between computer-aided diagnosis of breast masses and that of calcifications. Radiology 2002; 223: pp. 489-493.
26. Huo Z.M., Giger M.L., Vyborny C.J., et. al.: Breast cancer: effectiveness of computer-aided diagnosis—observer study with independent database of mammograms. Radiology 2002; 224: pp. 560-568.
27. Sahiner B., Chan H.P., Roubidoux M.A., et. al.: Computer-aided diagnosis of malignant and benign breast masses in 3d ultrasound volumes: effect on radiologists’ accuracy. Radiology 2007; 242: pp. 716-724.
28. Horsch K., Giger M.L., Vyborny C.J., et. al.: Classification of breast lesions with multimodality computer-aided diagnosis: observer study results on an independent clinical data set. Radiology 2006; 240: pp. 357-368.
29. Bhatti P.T., LeCarpentier G.L., Roubidoux M.A., et. al.: Discrimination of sonographically detected breast masses using frequency shift color Doppler imaging in combination with age and gray scale criteria. J Ultrasound Med 2001; 20: pp. 343-350.
30. Sahiner B., Petrick N., Chan H.P., et. al.: Computer-aided characterization of mammographic masses: accuracy of mass segmentation and its effects on characterization. IEEE Trans Med Imaging 2001; 20: pp. 1275-1284.
31. Hadjiiski L.M., Sahiner B., Chan H.P., et. al.: Analysis of temporal change of mammographic features: computer-aided classification of malignant and benign breast masses. Med Phys 2001; 28: pp. 2309-2317.
32. Sahiner B., Chan H.P., Petrick N., et. al.: Improvement of mammographic mass characterization using spiculation measures and morphological features. Medi Phys 2001; 28: pp. 1455-1465.
33. Sahiner B., Chan H.P., Petrick N., et. al.: Computerized characterization of masses on mammograms: the rubber band straightening transform and texture analysis. Med Phys 1998; 25: pp. 516-526.
34. Sahiner B., Chan H.P., Roubidoux M.A., et. al.: Computerized characterization of breast masses on 3-D ultrasound volumes. Med Phys 2004; 31: pp. 744-754.
35. American College of Radiology Breast Imaging Reporting and Data System Atlas (BI-RADS Atlas).2003.American College of RadiologyReston, VA
36. Sickles E.A.: Nonpalpable, circumscribed, noncalcified solid breast masses: likelihood of malignancy based on lesion size and age of patient. Radiology 1994; 192: pp. 439-442.
37. Metz C.E.: ROC methodology in radiologic imaging. Invest Radiol 1986; 21: pp. 720-733.
38. McNeil B.J., Hanley J.A.: Statistical approaches to the analysis of receiver operating characteristic (ROC) curves. Med Decis Making 1984; 4: pp. 137-150.
39. Dorfman D.D., Berbaum K.S., Metz C.E.: ROC rating analysis: generalization to the population of readers and cases with the jackknife method. Invest Radiol 1992; 27: pp. 723-731.
40. Jiang Y., Metz C.E., Nishikawa R.M.: A receiver operating characteristic partial area index for highly sensitive diagnostic tests. Radiology 1996; 201: pp. 745-750.
41. Hastie T., Tibshirani R., Friedman J.: The elements of statistical learning.2001.Springer-VerlagNew York
42. Sahiner B., Chan H.P., Hadjiiski L.: Classifier performance prediction for computer-aided diagnosis using a limited data set. Med Phys 2008; 35: pp. 1559-1570.
43. Gur D., Rockette H.E., Armfield D.R., et. al.: Prevalence effect in a laboratory environment. Radiology 2003; 228: pp. 10-14.
44. Gur D., Bandos A.I., Fuhrman C.R., et. al.: The prevalence effect in a laboratory environment: changing the confidence ratings. Acad Radiol 2007; 14: pp. 49-53.
45. Egglin T.K.P., Feinstein A.R.: Context bias–aproblem in diagnostic radiology. JAMA 1996; 276: pp. 1752-1755.
46. Gur D., Bandos A.I., Cohen C.S., et. al.: The “laboratory” effect: comparing radiologists’ performance and variability during prospective clinical and laboratory mammography interpretations. Radiology 2008; 249: pp. 47-53.