Rationale and Objective
This study aims to identify potential barriers to the clinical implementation of quantitative imaging for the assessment of tumor heterogeneity.
Materials and Methods
An 18-month prospective observational study was undertaken in which the clinical implementation of computed tomography texture analysis (CTTA) as a technique for quantifying tumor heterogeneity in patients with non–small cell lung cancer was assessed using the Reach, Effectiveness, Adoption, Implementation, and Maintenance (RE-AIM) framework.
Results
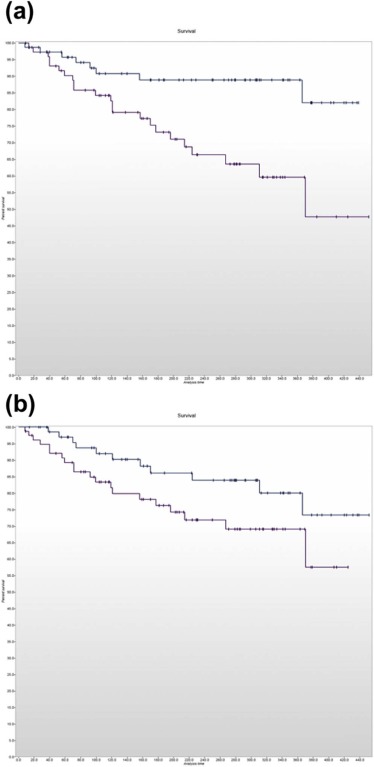
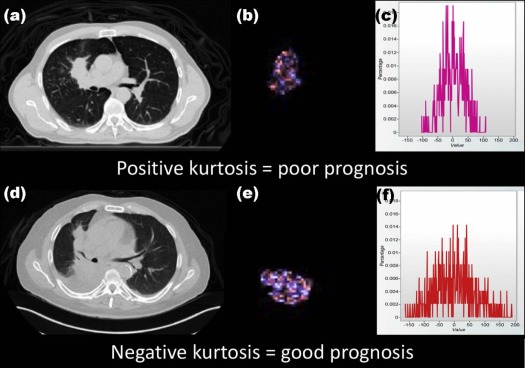
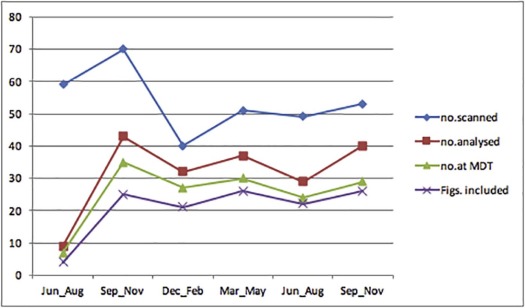
Adopters of the technology comprised five specialists with dual accreditation in radiology and nuclear medicine supervising two trainees. Tumor heterogeneity information was extracted and reported in 190 of 322 eligible cases (59%) and presented at the multidisciplinary team meeting in 124 of 152 patients (82%) for whom CTTA had been performed. The maximum proportion of eligible cases in which heterogeneity information had been extracted and reported in any quarter was 80%, but fell in the latter half of the study. The maximum frequency with which available CTTA results were presented at the multidisciplinary team meeting in any quarter was 92% and was maintained in the latter part of the study. Significant differences in survival were observed for patients categorized using the two reported CTTA values ( P = 0.004 and P = 0.0057, respectively).
Conclusions
Radiologist engagement is a potential barrier to the effective translation of quantitative imaging assessments of tumor heterogeneity into clinical practice and will need to be addressed before tumor heterogeneity information can successfully contribute to clinical decision making in oncology.
Introduction
Genomic and phenotypic heterogeneities are recognized features of malignancy that have prognostic significance and potential impact on treatment response . It is increasingly acknowledged that this biological heterogeneity is also represented within images produced by a range of routine diagnostic tests . A key aspect of tumor heterogeneity is the coexistence of genetically distinct subclones within a single tumor that is a consequence of underlying genetic instability . The ability of imaging to depict these subclones is reinforced by the cumulative identification of imaging correlates for a range of genomic aberrations in different tumor types , and by data from biopsy studies confirming the spatial separation of subclones to be sufficiently large for detection by imaging . Nevertheless, information reflecting tumor heterogeneity is rarely included in clinical reports from diagnostic imaging tests in oncology and, if included, is typically expressed in subjective descriptive terms rather than quantitative measures.
There is considerable potential for quantitative imaging of tumor heterogeneity to contribute to the clinical care of patients with cancer. Possible applications include the characterization of lesions as benign or malignant, for instance, in the evaluation of pulmonary nodules . For lesions known to be malignant, imaging measures of heterogeneity can potentially provide correlates for biological features such as gene mutations when tissue-based assays are contraindicated or have failed . Heterogeneity measures may also be useful in providing an indication of tumor aggression as illustrated by the association of heterogeneity within computed tomography (CT) images of primary lung cancers with the likelihood of mediastinal metastases and overall prognosis . Heterogeneity imaging can also potentially provide an indication of likely response to treatment. The application of quantifiable imaging characteristics as indices for disease status has a number of advantages over tissue-based assays. Imaging has an established role in cancer management, and tumor heterogeneity measurements can frequently be obtained as part of routine diagnosis. Imaging is noninvasive and can therefore be repeated at different stages during the evolution of the disease or treatment. Being closely related to the tumor phenotype, imaging assessments of tumor heterogeneity can provide information that is complementary to gene-based assays .
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and Methods
Study Design and Setting
Get Radiology Tree app to read full this article<
Patient Population and Imaging Technique
Get Radiology Tree app to read full this article<
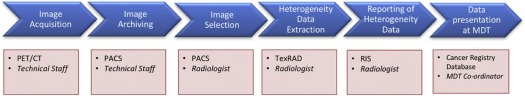
Clinical Workflow
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Stakeholder Engagement
Get Radiology Tree app to read full this article<
Outcome Measures
Get Radiology Tree app to read full this article<
Table 1
Summary of Outcome Measures for Each Reach, Effectiveness, Adoption, Implementation, and Maintenance Domain
Domains Outcome Measures Reach Proportion of eligible cases for which quantitative information on tumor heterogeneity was extracted and reported Effectiveness Comparison of the survival of patients with computed tomography texture analysis parameters above and below the prescribed threshold values Adoption The number and representativeness of radiologists extracting quantitative information on tumor heterogeneity from computed tomography Implementation Frequency with which available quantitative information on tumor heterogeneity is presented at the multidisciplinary team meeting Maintenance Change in reach, adoption, and implementation measures over time.
Get Radiology Tree app to read full this article<
Results
Reach
Get Radiology Tree app to read full this article<
Effectiveness
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Adoption
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Implementation
Get Radiology Tree app to read full this article<
Maintenance
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
Appendix 1A
Information Sheet for Radiologists
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Computed Tomography Texture Analysis (CTTA) in Non–small Cell Lung Cancer (NSCLC)
Information for Radiologists
The Aim of the Study:
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
The Process:
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion:
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Feedback and Follow-up:
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Appendix 1B
Information Sheet for Referring Clinicians
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Computed Tomography (CTTA) in Non–small Cell Lung Cancer (NSCLC)
Information for Clinicians
The Aim of the Study:
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
The Method:
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion:
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Feedback and Follow-up:
Get Radiology Tree app to read full this article<
Appendix 2
Quantitative Imaging for Tumor Heterogeneity Reporting Template
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Burrell R.A., McGranahan N., Bartek J., et. al.: The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013; 501: pp. 338. e45
2. Davnall F., Yip C.S.P., Ljungqvist G., et. al.: Assessment of tumor heterogeneity: an emerging imaging technique for clinical practice?. Insights Imaging 2012; 3: pp. 573-589.
3. O’Connor J.P.B., Rose C.J., Waterton J.C., et. al.: Imaging intratumor heterogeneity: role in therapy response, resistance and clinical outcome. Clin Cancer Res 2015; 21: pp. 249-257.
4. Bashir U., Siddique M.M., McLean E., et. al.: Imaging heterogeneity in lung cancer: techniques, applications and challenges. AJR Am J Radiol Roentgenol 2016; 207: pp. 534-543.
5. Cox V.L., Bhosale P., Varadhachary G.R., et. al.: Cancer genomics and important oncologic mutations: a contemporary guide for body imagers. Radiology 2017; 283: pp. 314-340.
6. Weiss G.J., Ganeshan B., Miles K.A., et. al.: Non-invasive image texture analysis differentiates K-ras mutation from pan-wildtype NSCLC and is prognostic. PLoS ONE 2014; 9: e100244
7. Caramella C., Bluthgen M.V., Rosellini S., et. al.: Prognostic value of texture analysis and correlation with molecular profile in EGFR mutated/ALK rearranged advanced non-small cell lung cancer (NSCLC). Europ J Cancer 2015; 51: pp. S647-S648.
8. Craigie M., Squires J., Miles K.: Can CT measures of tumour heterogeneity stratify risk for nodal metastasis in patients with non-small cell lung cancer?. Clin Radiol 2017; 72: pp. 899.e1-899.e7.
9. Win T., Miles K.A., Janes S.M., et. al.: Tumor heterogeneity and permeability as measured on the CT component of PET/CT predict survival in patients with non-small cell lung cancer. Clin Cancer Res 2013; 19: pp. 3591-3599.
10. Fried D.V., Tucker S.L., Zhou S., et. al.: Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int J Radiat Oncol Biol Phys 2014; 90: pp. 834-842.
11. Bluthgen M.V., Caramella C., Faire L., et. al.: Prognostic value of texture analysis in advanced non-small cell lung cancer (NSCLC). Europ J Cancer 2015; 51: pp. S645-S646.
12. Chowdhury R., Ganeshan B., Irshad S., et. al.: The use of molecular imaging combined with genomic techniques to understand the heterogeneity in cancer metastasis. Br J Radiol 2014; 8: pp. 20140065.
13. Lee C.I., Gupta S., Sherry S.J., et. al.: Translating new imaging technologies to clinical practice. Acad Radiol 2018; 25: pp. 3-8. Epub ahead of print 2017 August 31
14. Bakken S., Ruland C.M.: Translating clinical informatics interventions into routine clinical care: how can the RE-AIM framework help?. J Am Med Inform Assoc 2009; 16: pp. 889-897.
15. Miles K.A., Ganeshan B., Hayball M.P.: CT texture analysis using the filtration-histogram method: what do the measurements mean?. Cancer Imaging 2013; 13: pp. 400.
16. Miles K.A.: How to use CT texture analysis for prognostication of non-small cell lung cancer. Cancer Imaging 2016; 16: pp. 10.