Rationale and Objectives
This study aims to investigate the sensitivity of quantitative metrics derived from dynamic contrast-enhanced (DCE) magnetic resonance imaging and a pharmacokinetic (PK) model to image quality and acquisition parameters.
Materials and Methods
A computer-synthesized DCE model that consisted of a large range of values of K trans (transfer constant of a paramagnetic contrast agent from blood to tissue), v p (fractional plasma volume), and k ep (back flux rate) was created to test the reliability of quantitative metrics derived from a standard PK model. Effects of the contrast-to-noise ratio (CNR), total acquisition time, and sampling interval on the stability and bias of the derived metrics were investigated.
Results
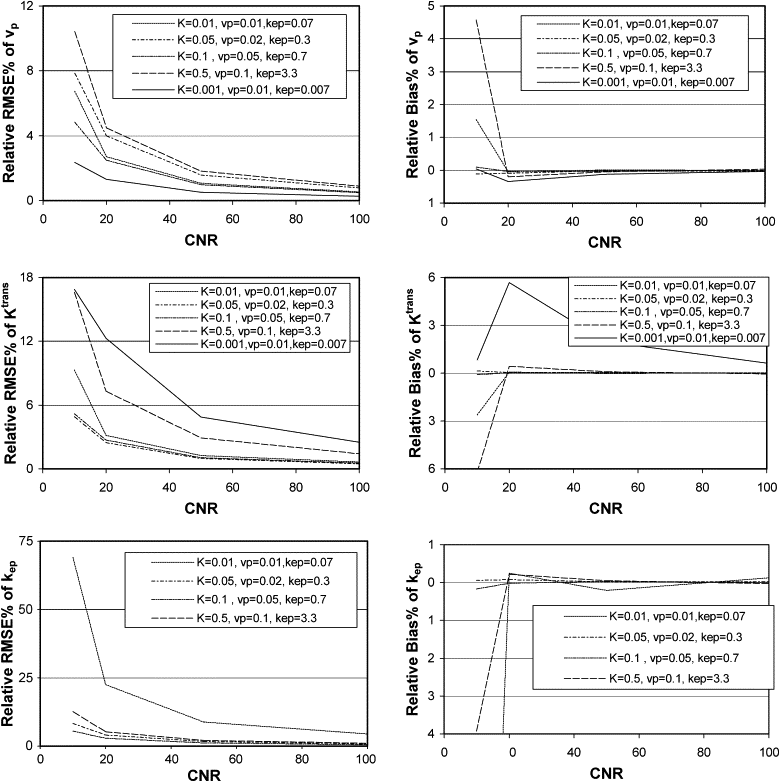
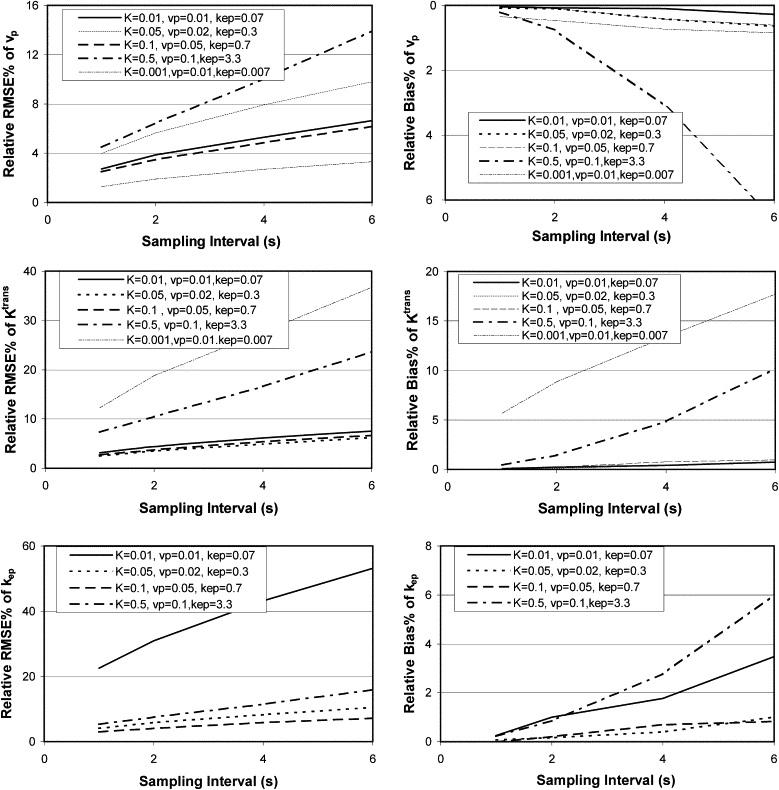
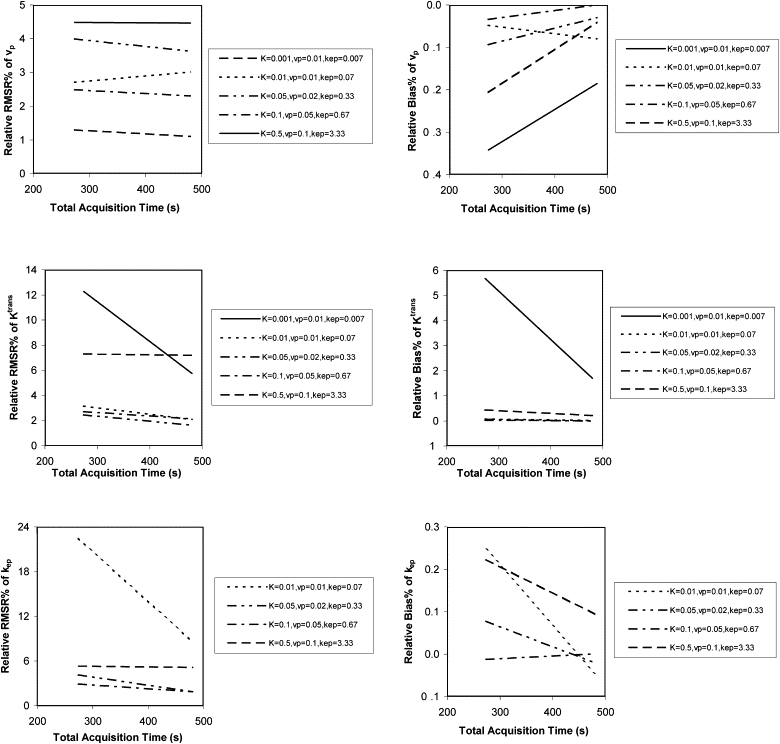
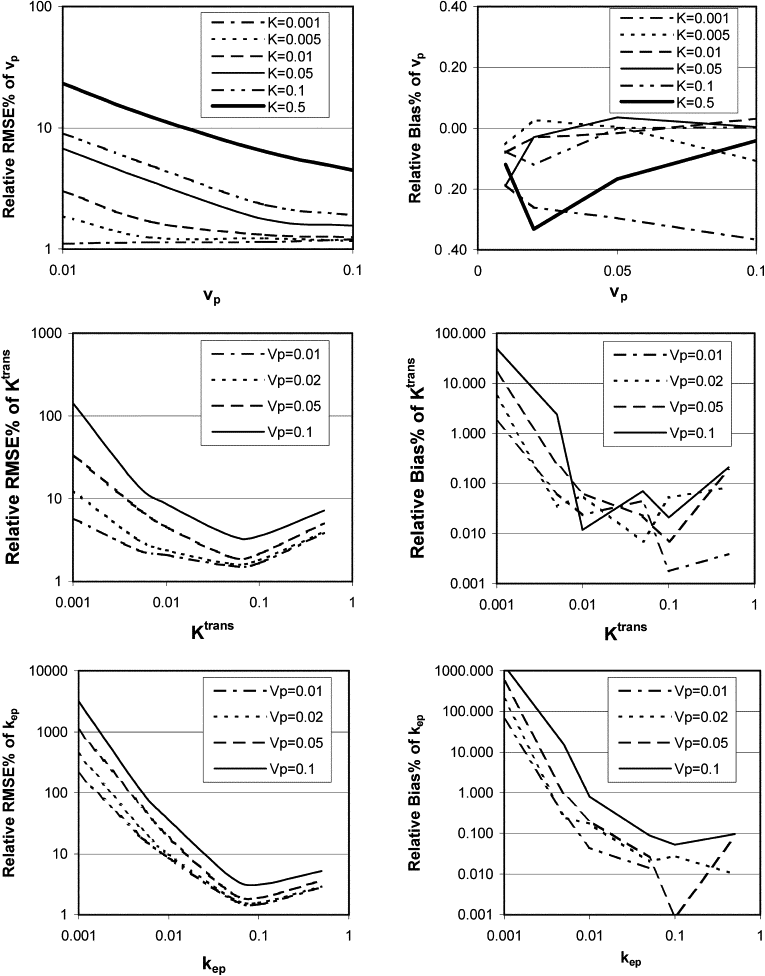
The instability and bias of the estimated K trans , v p , and k ep values increased with sampling interval and decreased with increasing CNR. Total acquisition times had limited influence on the estimations of K trans and v p values, but increasing the total acquisition time improved the stability of the estimation of k ep values. However, for small k ep values, the stability was still poor even with a total acquisition time of 8 minutes. Also, the stability and bias of the estimated values of K trans , v p , and k ep are interrelated.
Conclusions
Our synthesized DCE model represents perfectly reproduced data except for the presence of Gaussian-distributed random noise. Our analysis suggests minimum changes that may be considered potentially significant in longitudinal therapy assessment studies. Our data are complementary to experimental data from human subjects and phantoms, and provide guidance for the design of image acquisition strategies.
Dynamic contrast-enhanced (DCE) magnetic resonance (MRI) is emerging as a biomarker for assessment of cancer therapy and normal tissue toxicity . Quantitative metrics obtained by applying a pharmacokinetic (PK) model to DCE MRI have been related to underlying physiological parameters such as vascular permeability, vascular volume, perfusion, and extravascular extracellular space. Previous studies have investigated factors influencing accuracy and precision of quantitative metrics derived from DCE MRI and the PK models. Most attention has been given to the factors that impact measurement of the arterial input function, such as the amplitude, sampling rate, temporal jitter, dispersion, partial volume, and inflow effect, as well as the propagation of these measurement errors on the derived quantitative metrics . However, few studies address reproducibility of DCE MRI measures and stability of the PK model, both of which affect reliability of subsequently derived quantitative metrics and validity of the metrics as a biomarker for therapy assessment.
Recently, efforts have been made to standardize DCE MRI acquisition protocols, establish quality assurance procedures for multicenter clinical trials, and assess reproducibility of test and retest measures. Although the importance of reproducibility of DCE MRI measures as well as the derived quantitative metrics is well recognized, there are few published data to provide guidelines for investigators . Furthermore, reproducibility of the quantitative metrics derived from a PK model and DCE MRI depends on many interconnected factors, including stability of scanner hardware, physiological conditions, image acquisition parameters, image quality, and the analysis method used. Therefore, it is important to understand the impact of physiological-related, measurement-related, as well as quantification method–related effects on reproducibility of the quantitative metrics, and to develop strategies to improve reproducibility while considering these effects. Few studies to date have investigated the stability of the PK model and associated computational algorithms under the influence of image acquisition parameters and noise .
Get Radiology Tree app to read full this article<
Materials and methods
Standard PK Model
Get Radiology Tree app to read full this article<
Ct(t)Ktrans∫otCp(τ)e−Kep(t−τ)dτ+vpCp(t) C
t
(
t
)
K
t
r
a
n
s
∫
o
t
C
p
(
τ
)
e
−
K
e
p
(
t
−
τ
)
d
τ
+
v
p
C
p
(
t
)
where C t and C p are the contrast concentrations in tissue and arterial plasma, respectively, K trans is the transfer constant of a paramagnetic contrast agent from intravascular space to extravascular space, v p is the fractional plasma volume, κ ep = K trans /v e depicts the back flux rate of the contrast agent from tissue to plasma, and v e is the fractional volume of extravascular extracellular space. Given a set of DCE MRI data, three parameters, K trans , v p , and v e , can be estimated from Equation 1 . The precise relationship of these quantitative metrics ( K trans , v p , and k ep [or v e ]) to true underlying physiological parameters is hardly validated histopathologically . The utility of the measurements depends on physiological conditions under study , and interpretation of extracted quantitative metrics should be handled with caution. In this project, we investigated the sensitivity of K trans , v p , and k ep (or v e ) as derived from DCE imaging data using Equation 1 with the CNR, sampling interval, and total acquisition time.
Get Radiology Tree app to read full this article<
Synthesized DCE Phantom
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Testing of the Synthesized Phantom
Get Radiology Tree app to read full this article<
Results
Validation of PK Modeling Software
Get Radiology Tree app to read full this article<
Stability and Bias of Derived Quantitative Metrics
CNR
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Sampling interval
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Total acquisition time
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Interaction among metrics selves
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
95% confidence intervals
Get Radiology Tree app to read full this article<
Table 1
95% Confidence Intervals at CNR = 20, Δt = 1second, and Total Acquisition Time = 274 seconds
v p__K trans__k ep True Value 95% CI 1.96 RMSE/M (%) True Value 95% CI 1.96 RMSE/M (%) True Value 95% CI 1.96 RMSE/M (%) 0.01 0.0097 0.0102 2.6 0.001 0.0008 0.0013 22.8 0.0067 −0.0669 0.1297 313.0 0.01 0.0097 0.0103 3.1 0.005 0.0047 0.0054 7.0 0.0333 0.0004 0.0660 98.8 0.01 0.0095 0.0105 5.3 0.010 0.0094 0.0106 6.2 0.0667 0.0374 0.0962 44.0 0.01 0.0085 0.0114 14.5 0.050 0.0478 0.0523 4.5 0.3333 0.3079 0.3591 7.7 0.01 0.0081 0.0119 19.3 0.100 0.0960 0.1042 4.1 0.6667 0.6374 0.6967 4.4 0.01 0.0054 0.0146 45.7 0.500 0.4632 0.5372 7.4 3.3333 3.1538 3.5156 5.4 0.02 0.0194 0.0204 2.5 0.001 0.0006 0.0016 45.3 0.0067 −0.1347 0.2552 323.7 0.02 0.0194 0.0205 2.8 0.005 0.0044 0.0056 11.9 0.0333 −0.0177 0.0905 148.6 0.02 0.0194 0.0206 3.1 0.010 0.0093 0.0107 7.4 0.0667 0.0319 0.1017 52.2 0.02 0.0184 0.0215 7.8 0.050 0.0476 0.0524 4.8 0.3333 0.3065 0.3606 8.1 0.02 0.0180 0.0220 10.1 0.100 0.0958 0.1043 4.3 0.6667 0.6357 0.6978 4.7 0.02 0.0151 0.0248 24.2 0.500 0.4609 0.5398 7.9 3.3333 3.1408 3.5281 5.8 0.05 0.0485 0.0511 2.7 0.001 −0.0006 0.0035 144.0 0.0067 −0.4604 0.7826 385.8 0.05 0.0486 0.0512 2.6 0.005 0.0039 0.0066 25.9 0.0333 −0.0641 0.1650 227.0 0.05 0.0485 0.0515 2.9 0.010 0.0084 0.0117 16.6 0.0667 −0.0102 0.1468 114.9 0.05 0.0482 0.0518 3.6 0.050 0.0471 0.0529 5.7 0.3333 0.3001 0.3662 9.9 0.05 0.0475 0.0524 4.9 0.100 0.0947 0.1053 5.3 0.6667 0.6282 0.7049 5.8 0.05 0.0441 0.0561 12.0 0.500 0.4502 0.5486 9.9 3.3333 3.0863 3.5724 7.3 0.10 0.0939 0.1046 5.4 0.001 −0.3580 0.3746 4394.8 0.0067 −5.3816 6.5883 992.0 0.10 0.0970 0.1023 2.7 0.005 0.0027 0.0085 51.1 0.0333 −0.1424 0.3048 275.4 0.10 0.0972 0.1026 2.7 0.010 0.0073 0.0132 29.1 0.0667 −0.0573 0.2085 175.9 0.10 0.0966 0.1033 3.4 0.050 0.0448 0.0552 10.4 0.3333 0.2729 0.3926 18.0 0.10 0.0961 0.1039 3.9 0.100 0.0916 0.1086 8.5 0.6667 0.6053 0.7295 9.3 0.10 0.0910 0.1086 8.8 0.500 0.4306 0.5738 14.3 3.3333 2.9952 3.6863 10.3
CNR, contrast-to-noise ratio; k ep , back flux rate; K trans , transfer constant of a paramagnetic contrast agent from blood to tissue; RMSE, root mean squared error/mean; v p , fractional plasma volume.
95% CI: mean ± 1.96 ∗ RMSE for single estimation (eg, single voxel estimation); 1.96 RMSE/M (%): 100 ∗ 1.96 ∗ RMSE/mean.
Table 2
95% Confidence Intervals at CNR = 20, Δt = 4 seconds, and Total Acquisition Time = 274 seconds
v p__K trans__k ep True Value 95% CI 1.96 RMSE/M (%) True Value 95% CI 1.96 RMSE/M (%) True Value 95% CI 1.96 RMSE/M (%) 0.01 0.009 0.010 5.3 0.001 0.0006 0.0017 47.7 0.0067 −0.1416 0.2628 333.6 0.01 0.009 0.011 5.9 0.005 0.0044 0.0057 12.6 0.0333 −0.0216 0.0942 159.6 0.01 0.009 0.011 10.4 0.010 0.0088 0.0112 12.0 0.0667 0.0114 0.1243 83.2 0.01 0.007 0.013 29.3 0.050 0.0458 0.0548 8.9 0.3333 0.2858 0.3865 15.0 0.01 0.006 0.014 41.2 0.100 0.0920 0.1094 8.6 0.6667 0.6093 0.7325 9.2 0.01 −0.003 0.018 141.3 0.500 0.4342 0.6034 16.3 3.3333 3.0090 3.8116 11.8 0.02 0.019 0.021 5.4 0.001 0.0000 0.0027 102.2 0.0067 −0.3211 0.5755 352.4 0.02 0.019 0.021 5.6 0.005 0.0040 0.0063 22.3 0.0333 −0.0513 0.1430 211.8 0.02 0.019 0.021 6.2 0.010 0.0086 0.0115 14.4 0.0667 0.0008 0.1359 98.8 0.02 0.017 0.023 15.6 0.050 0.0454 0.0550 9.6 0.3333 0.2807 0.3887 16.1 0.02 0.016 0.024 21.0 0.100 0.0916 0.1097 9.0 0.6667 0.6059 0.7351 9.6 0.02 0.005 0.029 68.4 0.500 0.4266 0.6155 18.1 3.3333 2.9806 3.8543 12.8 0.05 0.043 0.056 12.8 0.001 −0.1372 0.1528 1848.7 0.0067 −4.4544 5.6988 815.9 0.05 0.047 0.052 5.4 0.005 0.0026 0.0087 53.8 0.0333 −0.1492 0.3195 275.1 0.05 0.047 0.053 5.7 0.010 0.0073 0.0134 29.7 0.0667 −0.0560 0.2122 171.7 0.05 0.046 0.054 7.3 0.050 0.0445 0.0561 11.6 0.3333 0.2699 0.4018 19.6 0.05 0.045 0.055 9.6 0.100 0.0901 0.1114 10.6 0.6667 0.5942 0.7484 11.5 0.05 0.033 0.061 29.6 0.500 0.4049 0.6343 22.1 3.3333 2.8710 3.9480 15.8 0.10 0.077 0.118 20.8 0.001 −0.4600 0.5258 1498.0 0.0067 −7.2555 9.4160 771.6 0.10 0.094 0.105 5.4 0.005 −0.0023 0.0157 134.1 0.0333 −0.3802 0.6799 353.7 0.10 0.094 0.105 5.3 0.010 0.0049 0.0170 55.3 0.0667 −0.1401 0.3485 234.5 0.10 0.093 0.107 6.8 0.050 0.0394 0.0611 21.5 0.3333 0.2121 0.4551 36.4 0.10 0.092 0.108 7.9 0.100 0.0840 0.1178 16.7 0.6667 0.5517 0.7922 17.9 0.10 0.077 0.117 20.3 0.500 0.3615 0.6871 31.1 3.3333 2.6716 4.1801 22.0
CNR, contrast-to-noise ratio; k ep , back flux rate; K trans , transfer constant of a paramagnetic contrast agent from blood to tissue; RMSE, root mean squared error/mean; v p , fractional plasma volume.
95% CI: mean ± 1.96 ∗ RMSE for single estimation (eg, single voxel estimation); 1.96 RMSE/M (%): 100 ∗ 1.96 ∗ RMSE/mean.
Table 3
95% Confidence Intervals at CNR = 20, Δt = 1second, and Total Acquisition Time = 480 seconds
v p__K trans__k ep True value 95% CI 1.96 RMSE/M (%) True value 95% CI 1.96 RMSE/M (%) True value 95% CI 1.96 RMSE/M (%) 0.01 0.0098 0.0102 2.2 0.001 0.0009 0.0011 11.1 0.0067 −0.0157 0.0373 244.8 0.01 0.0096 0.0104 3.7 0.005 0.0048 0.0052 4.7 0.0333 0.0213 0.0455 36.2 0.01 0.0094 0.0106 5.9 0.010 0.0096 0.0104 4.1 0.0667 0.0555 0.0778 16.7 0.01 0.0087 0.0113 13.2 0.050 0.0485 0.0515 3.0 0.3333 0.3217 0.3451 3.5 0.01 0.0082 0.0118 17.6 0.100 0.0968 0.1033 3.2 0.6667 0.6475 0.6857 2.9 0.01 0.0055 0.0145 45.4 0.500 0.4622 0.5379 7.6 3.3333 3.1500 3.5138 5.5 0.02 0.0195 0.0204 2.2 0.001 0.0008 0.0013 23.3 0.0067 −0.0362 0.0762 280.7 0.02 0.0195 0.0205 2.4 0.005 0.0047 0.0053 6.2 0.0333 0.0169 0.0500 49.5 0.02 0.0193 0.0207 3.3 0.010 0.0095 0.0105 4.7 0.0667 0.0540 0.0795 19.1 0.02 0.0186 0.0214 7.1 0.050 0.0484 0.0516 3.2 0.3333 0.3210 0.3455 3.7 0.02 0.0181 0.0219 9.6 0.100 0.0965 0.1036 3.5 0.6667 0.6465 0.6872 3.1 0.02 0.0151 0.0248 24.4 0.500 0.4613 0.5396 7.8 3.3333 3.1453 3.5221 5.7 0.05 0.0487 0.0510 2.2 0.001 0.0005 0.0018 57.3 0.0067 −0.0949 0.1806 321.4 0.05 0.0488 0.0512 2.4 0.005 0.0043 0.0058 15.0 0.0333 −0.0053 0.0726 115.9 0.05 0.0487 0.0513 2.6 0.010 0.0091 0.0109 8.9 0.0667 0.0425 0.0905 36.1 0.05 0.0483 0.0517 3.4 0.050 0.0481 0.0519 3.9 0.3333 0.3185 0.3480 4.4 0.05 0.0477 0.0523 4.5 0.100 0.0958 0.1042 4.2 0.6667 0.6422 0.6911 3.7 0.05 0.0437 0.0561 12.5 0.500 0.4514 0.5504 9.9 3.3333 3.1026 3.5693 7.0 0.10 0.0973 0.1020 2.4 0.001 −0.0013 0.0043 187.5 0.0067 −0.3164 0.5093 428.0 0.10 0.0976 0.1022 2.3 0.005 0.0037 0.0065 27.4 0.0333 −0.0305 0.1073 179.3 0.10 0.0976 0.1025 2.5 0.010 0.0083 0.0117 16.9 0.0667 0.0198 0.1125 70.1 0.10 0.0970 0.1030 3.0 0.050 0.0466 0.0535 6.9 0.3333 0.3074 0.3599 7.9 0.10 0.0963 0.1037 3.7 0.100 0.0931 0.1069 6.9 0.6667 0.6261 0.7065 6.0 0.10 0.0912 0.1087 8.7 0.500 0.4305 0.5716 14.1 3.3333 3.0002 3.6728 10.1
CNR, contrast-to-noise ratio; k ep , back flux rate; K trans , transfer constant of a paramagnetic contrast agent from blood to tissue; RMSE, root mean squared error/mean; v p , fractional plasma volume.
95% CI: mean ± 1.96 ∗ RMSE for single estimation (eg, single voxel estimation); 1.96 RMSE/M (%): 100 ∗ 1.96 ∗ RMSE/mean that defines a relative 95% confidence interval.
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Galbraith S.M., Rustin G.J., Lodge M.A., et. al.: Effects of 5,6-dimethylxanthenone-4-acetic acid on human tumor microcirculation assessed by dynamic contrast-enhanced magnetic resonance imaging. J Clin Oncol 2002; 20: pp. 3826-3840.
2. Morgan B., Thomas A.L., Drevs J., et. al.: Dynamic contrast-enhanced magnetic resonance imaging as a biomarker for the pharmacological response of PTK787/ZK 222584, an inhibitor of the vascular endothelial growth factor receptor tyrosine kinases, in patients with advanced colorectal cancer and liver metastases: results from two phase I studies. J Clin Oncol 2003; 21: pp. 3955-3964.
3. Rugo H.S., Herbst R.S., Liu G., et. al.: Phase I trial of the oral antiangiogenesis agent AG-013736 in patients with advanced solid tumors: pharmacokinetic and clinical results. J Clin Oncol 2005; 23: pp. 5474-5483.
4. Sorensen A.G., Batchelor T.T., Zhang W.T., et. al.: A “vascular normalization index” as potential mechanistic biomarker to predict survival after a single dose of cediranib in recurrent glioblastoma patients. Cancer Res 2009; 69: pp. 5296-5300.
5. Cao Y., Popovtzer A., Li D., et. al.: Early prediction of outcome in advanced head-and-neck cancer based on tumor blood volume alterations during therapy: a prospective study. Int J Radiat Oncol Biol Phys 2008; 72: pp. 1287-1290.
6. Cao Y., Tsien C.I., Sundgren P., et. al.: DCE MRI as a biomarker for prediction of radiation-induced neurocognitive dysfunction. Clin Cancer Res 2009; 15: pp. 1747-1754.
7. Henderson E., Rutt B.K., Lee T.Y.: Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magn Reson Imaging 1998; 16: pp. 1057-1073.
8. Calamante F., Gadian D.G., Connelly A.: Delay and dispersion effects in dynamic susceptibility contrast MRI: simulations using singular value decomposition. Magn Reson Med 2000; 44: pp. 466-473.
9. Cheng H.L.: Investigation and optimization of parameter accuracy in dynamic contrast-enhanced MRI. J Magn Reson Imaging 2008; 28: pp. 736-743.
10. Peeters F., Annet L., Hermoye L., Van Beers B.E.: Inflow correction of hepatic perfusion measurements using T1-weighted, fast gradient-echo, contrast-enhanced MRI. Magn Reson Med 2004; 51: pp. 710-717.
11. Singh A., Rathore R.K., Haris M., et. al.: Improved bolus arrival time and arterial input function estimation for tracer kinetic analysis in DCE-MRI. J Magn Reson Imaging 2009; 29: pp. 166-176.
12. Jackson A., Jayson G.C., Li K.L., et. al.: Reproducibility of quantitative dynamic contrast-enhanced MRI in newly presenting glioma. Br J Radiol 2003; 76: pp. 153-162.
13. Galbraith S.M., Lodge M.A., Taylor N.J., et. al.: Reproducibility of dynamic contrast-enhanced MRI in human muscle and tumours: comparison of quantitative and semi-quantitative analysis. NMR Biomed 2002; 15: pp. 132-142.
14. Padhani A.R., Hayes C., Landau S., et. al.: Reproducibility of quantitative dynamic MRI of normal human tissues. NMR Biomed 2002; 15: pp. 143-153.
15. Yang C., Karczmar G.S., Medved M., et. al.: Reproducibility assessment of a multiple reference tissue method for quantitative dynamic contrast enhanced-MRI analysis. Magn Reson Med 2009; 61: pp. 851-859.
16. Lopata R.G., Backes W.H., van den Bosch P.P., et. al.: On the identifiability of pharmacokinetic parameters in dynamic contrast-enhanced imaging. Magn Reson Med 2007; 58: pp. 425-429.
17. Tofts P.S., Brix G., Buckley D.L., et. al.: Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 1999; 10: pp. 223-232.
18. Ewing J.R., Knight R.A., Nagaraja T.N., et. al.: Patlak plots of Gd-DTPA MRI data yield blood-brain transfer constants concordant with those of 14C-sucrose in areas of blood-brain opening. Magn Reson in Med 2003; 50: pp. 283-292.
19. Cao Y.: Development of Image Software Tools for Radiation Therapy Assessment. Medical Physics 2005; 32: pp. 2136.