Rationale and Objectives
This article provides a survey of segmentation methods for medical images. Usually, classification of segmentation methods is done based on the approaches adopted and the domain of application.
Materials and Methods
This survey is conducted on the recent segmentation methods used in biomedical image processing and explores the methods useful for better segmentation. A critical appraisal of the current status of semiautomated and automated methods is made for the segmentation of anatomical medical images emphasizing the advantages and disadvantages. Computer-aided diagnosis (CAD) used by radiologists as a second opinion has become one of the major research areas in medical imaging and diagnostic radiology. A picture archiving communication system (PACS) is an integrated workflow system for managing images and related data that is designed to streamline operations throughout the whole patient care delivery process.
Results
By using PACS, the medical image interpretation may be changed from conventional hard-copy images to soft-copy studies viewed on the systems workstations.
Conclusion
The automatic segmentations assist the doctors in making quick diagnosis. The CAD need not be comparable to that of physicians, but is surely complementary.
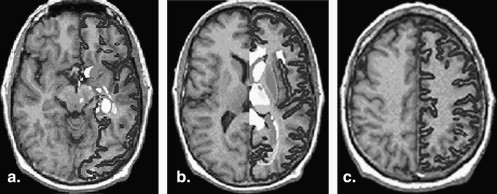
Tumor segmentation from magnetic resonance imaging (MRI) data is an important but time-consuming manual task performed by medical experts. Automating this process is a challenging task because of the high diversity in the appearance of tumor tissues among different patients and in many cases similarity with the normal tissues. MRI is an advanced medical imaging technique providing rich information about the human soft-tissue anatomy. It has several advantages over other imaging techniques, enabling it to provide three-dimensional data with high contrast between soft tissues. However, the amount of data is far too much for manual analysis, which has been one of the biggest obstacles in the effective use of MRI. For this reason, automatic or semiautomatic techniques of computer-aided image analysis are necessary. Segmentation of MRIs into different tissue classes, especially gray matter, white matter, and cerebrospinal fluid, is an important task. Brain MRIs have a number of features: first, they are statistically simple and are theoretically piecewise constant with a small number of classes. Second, they have relatively high contrast between different tissues. Unlike many other medical imaging modalities, the contrast in an MRI depends strongly on the way the image is acquired. By adding radio frequency or gradient pulses and by carefully choosing relaxation timings, it is possible to highlight different components in the object being imaged and produce high-contrast images. These two features facilitate segmentation. On the other hand, ideal imaging conditions are never realized in practice. The piecewise-constant property is degraded considerably by electronic noise, the bias field, and the partial-volume effect, all of which cause classes to overlap in the image intensity histogram. A wide variety of approaches have been proposed for brain MRI segmentation. These can be roughly divided into two categories: structural and statistical. Structural methods are based on the spatial properties of the image such as edges and regions. Various edge detection algorithms have been applied to extract boundaries between different brain tissues . However, such algorithms are vulnerable to artifacts and noise. Region growing is another popular structural approach. Starting from a totally different viewpoint, statistical methods label pixels according to probability values, which are determined based on the intensity distribution of the image. In their simplest form, thresholding-based methods are always chosen for scenes containing solid objects, resting on a background with intensities well separated from the objects. However, this is generally not effective for brain MRIs. Therefore, thresholding-based methods are unlikely to produce reliable results . Most statistical approaches rely on certain assumptions or models of the probability distribution function of the image intensities and its associated class labels, which can both be considered random variables. A statistical approach can either be parametric or nonparametric. Both are widely used in segmentation of brain MRIs. In nonparametric methods, the density model relies entirely on the data itself (ie, no prior assumption is made about the functional form of the distribution, but a large number of correctly labeled training points are required in advance). One of the most widely used nonparametric methods is K-Nearest-Neighbors. Nonparametric methods are adaptive, but suffer from the difficulty of obtaining a large number of training points, which can be tedious and a heavy burden even for experienced people. Clearly, such methods are not fully automatic. Unlike nonparametric approaches, parametric approaches rely on an explicit functional form of the intensity density function. For brain MRIs, the only method developed to date is based on the finite mixture model; in particular, the finite Gaussian mixture model when the Gaussian likelihood distribution is assumed .
Computer-aided diagnosis (CAD) has become a part of the routine clinical work for the detection of breast cancer on mammograms at many screening sites and hospitals in the United States. This indicates that CAD is beginning to be applied widely in the detection and differential diagnosis of many different types of abnormalities in medical images obtained in various examinations by use of different imaging modalities. In fact, CAD has become one of the major research subjects in medical imaging and diagnostic radiology . Although early attempts at computerized analysis of medical images were made in the 1960s, serious and systematic investigation on CAD began in the 1980s with a fundamental change in the concept for utilization of the computer output from automated computer diagnosis to CAD. Here, the motivation for development of CAD schemes is presented together with the current status and future potential of CAD in the environment of picture archiving communication system (PACS) . With the increasing availability of digitized images, CAD is a hot topic of research today. The basic concept is to provide a computer output as a second opinion to assist image interpretation by radiologists toward improving the accuracy and consistency of radiological diagnosis and also by reducing the image reading time. To achieve this goal, it is necessary not only to develop suitable algorithms, but also to quantify and maximize the effect of computer output on the performance of radiologists. Research and development of CAD involve a team effort by investigators–physicists, radiologists, computer scientists, engineers, psychologists, and statisticians. Use of all imaging modalities, all body parts and all kinds of examination, will have a major impact on medical imaging and diagnostic radiology .
Get Radiology Tree app to read full this article<
Literature review and research motivation
Get Radiology Tree app to read full this article<
Table 1
Summary of Clinical Evaluation Setup and Percentage Errors for Volume Measurement
Data Galima LG Galima HG Year Volume Error (%) Computation Time (Minutes) Gibbs T1E 10 1996 5 10 Letteboer 20 2004 8 — Droske T1E — 2005 Good 3 Liu FLAIR, T1, T1E 10 2005516 Vaidyanathan T1, PD, T2 4 199512 — Fletcher-Heath T1, PD, T2 6 200128 — Clark T1, PD, T2 (all with Gd) 7 19985 — Kaus SPGR-Enh 14 2001175 Moonis FLAIR 19 200117 — Mazzra T1, FLAIR (CT) 3 8 20041575 Prastawa T2 1 1 20042690
Galima LG, low grade galima; Galima HG, high grade galima; T1, longitudinal relaxation time; T2, transverse relaxation time; T1E, echo delay time; SPGR-Enh, Spoiled gradient-recalled-Enhanced; PD, proton-density; FLAIR, fluid-attenuated inversion-recovery; CT, computed tomography; Gd, Godolinium.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Classification of segmentation methods
Get Radiology Tree app to read full this article<
Thresholding
Get Radiology Tree app to read full this article<
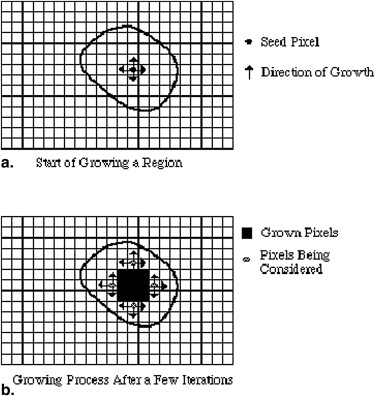
Region Growing
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Classifiers
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Clustering
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Artificial Neural Networks
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Markov Random Field Models
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Deformable Models
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Atlas-guided Approaches
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
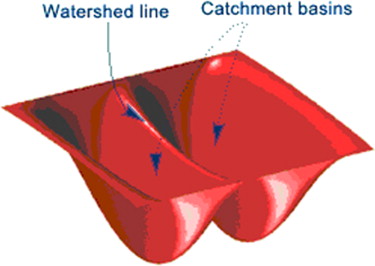
Watershed Methods
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Level Set Methods
Get Radiology Tree app to read full this article<
The Combination of Watershed and Level Set
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Fuzzy Connectedness
Get Radiology Tree app to read full this article<
Some Current Hot Techniques
The core neurochemical and imaging biomarkers of Alzheimer’s disease
Get Radiology Tree app to read full this article<
MRI-based volumetry
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Multiparametric tissue characterization of brain neoplasms using MRI
Get Radiology Tree app to read full this article<
Computer-assisted segmentation of white matter lesions in 3D MRI
Get Radiology Tree app to read full this article<
Segmentation of lung nodules in CT image
Get Radiology Tree app to read full this article<
Lung motion and volume measurements by dynamic 3D MRI
Get Radiology Tree app to read full this article<
Molecular positron emission tomography (PET)/CT imaging guided radiation therapy treatment planning
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Deformation registration of endorectal prostate MRI
Get Radiology Tree app to read full this article<
Detection of asthma using of multidetector CT and hyperpolarized He-3 MRI
Get Radiology Tree app to read full this article<
Automatic identification of infarct slices and hemisphere in diffusion-weighted imaging (DWI) scans
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Automated 11 C-PiB standardized uptake value ratio
Get Radiology Tree app to read full this article<
The current diagnosis packages
The Status of CAD
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 2
Number of Computed-assisted Diagnosis Papers Presented at the Radiological Society of North America 2000–2005
Years 2000 2001 2002 2003 2004 2005 Chest 22 37 53 94 70 48 Breast 23 28 32 37 48 49 Colon 4 10 21 17 15 30 Brain — 4 2 10 9 15 Liver 3 — 5 9 9 9 Skeletal 2 7 7 9 8 5 Vascular, etc 5 — 12 15 2 7 Total 59 86 134 191 161 163
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
The Status of PACS
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Table 3
Picture Archiving Communication System Usage in Europe
Study Number Country Approximate % of Picture Archiving Communication System Usage in Europe 1 France 10 2 Poland 11 3 Lithuania 20 4 Latvia 28 5 Germany 28 6 Greece 31 7 Czech Republic 33 8 Sweden 38 9 UK 41 10 Turkey 42 11 Spain 43 12 Portugal 43 13 Italy 48 14 Netherlands 50 15 Belgium 55 16 Hungary 55 17 Norway 67 18 Finland 69.5
Get Radiology Tree app to read full this article<
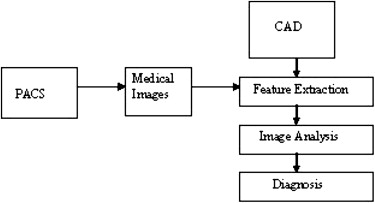
The role of CAD in the PACS environment
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Bomans M., Hohne K.H., Kramer L.H., et. al.: 3-D segmentation of MR images of the head for 3-D display. IEEE Trans Med Imaging 1990; 18: pp. 25-34.
2. Dellepiane S.: Image segmentation: errors, sensitivity, and uncertainty. Proc IEEE-Eng Med Biol Soc 1991; 13: pp. 253-254.
3. Clarke L.P., Velthuizen R.P., Camacho M.A., et. al.: MRI segmentation: methods and applications. Magnet Reson Imaging 1995; 13: pp. 343-368.
4. Brice C.R., Fennema C.L.: Scene analysis using regions. Artif Intell 1970; 1: pp. 205-226.
5. Zhu S.C., Yuille A.: Region competition: unifying snakes, region growing, and Bayes/MDL for multiband image segmentation. IEEE Trans Patt Anal Machine Intell 1996; 18: pp. 884-900.
6. Lim K.O., Pfefferbaum A.: Segmentation of MR brain images into cerebrospinal fluid spaces, with and gray matter. J Comp Assisted Tomogr 1989; 13: pp. 588-593.
7. Brummer M.E.: Optimized intensity thresholds for volumetric analysis of magnetic resonance imaging data. Pro SPIE 1808; 1992: pp. 299-310.
8. Kundu A.: Local segmentation of biomedical images. Comp Med Imaging Graph 1990; 14: pp. 173-183.
9. Wells W.M., Grimson E.L., Kikinis R., et. al.: Adaptive segmentation of MRI data. IEEE Trans Med Imaging 1996; 15: pp. 429-442.
10. Guillemaud R., Brady J.M.: Estimating the bias field of MR images. IEEE Trans Med Imaging 1997; 16: pp. 238-251.
11. Held K., Kops E.R., Krause B.J., et. al.: Markov random field segmentation of brain MR images. IEEE Trans Med Imaging 1997; 16: pp. 878-886.
12. Freer T.W., Ulissey M.J.: Screening mammography with computer-aided detection: prospective study of 12,860 patients in a community breast center. Radiology 2001; 220: pp. 781-786.
13. Gur D., Sumkin J.H., Rockette H.E., et. al.: Changes in breast cancer detection and mammography recall rate after the introduction of a computer-aided detection system. J Natl Cancer Inst 2004; 96: pp. 185-190.
14. Birdwell R.L., Bandodkar P., Ikeda D.M.: Computer-aided detection with screening mammography in a university hospital setting. Radiology 2005; 236: pp. 451-457.
15. Cupples T.E., Cunningham J.E., Reynolds J.C.: Impact of computer-aided detection in a regional screening mammography program. AJR Am J Roentgenol 2005; 185: pp. 944-950.
16. Morton M.J., Whaley D.H., Brandt K.R., et. al.: Screening mammograms: interpretation with computer-aided detection-prospective evaluation. Radiology 2006; 239: pp. 375-383.
17. Dean J.C., Ilvento C.C.: Improved cancer detection using computer-aided detection with diagnostic and screening mammography: prospective study of 104 cancers. AJR Am J Roentgenol 2006; 187: pp. 20-28.
18. Destounis S.V., DiNitto P., Logan-Young W., et. al.: Can computer-aided detection with double reading of screening mammograms help decrease the false-negative rate?. Radiology 2004; 232: pp. 578-584.
19. Butler S.A., Gabbay R.J., Kass D.A., et. al.: Computer-aided detection in diagnostic mammography: detection of clinically unsuspected cancers. AJR Am J Roentgenol 2004; 183: pp. 1511-1515.
20. Nishikawa R.M., Haldemann R.C., Papaioannou J., et. al.: Initial experience with a prototype clinical “intelligent” mammography workstation for computer-aided diagnosis. Proc SPIE 1995; 2434: pp. 65-71.
21. Schmidt R.A., Nishikawa R.M., Osnis R.B., et. al.: Computerized detection of lesions missed by mammography.1996.Elsevier ScienceAmsterdam
22. Warren-Burhenne L.J., Wood S.A., Orsi D., et. al.: Potential contribution of computer-aided detection to the sensitivity of screening mammography. Radiology 2000; 215: pp. 554-562.
23. Giger M.L., Huo Z., Kupinski M.A., et. al.: Computer-aided diagnosis in mammography. The handbook of medical imaging and medical imaging processing and analysis. SPIE 2000; 2: pp. 915-1004.
24. Giger ML. Computerized analysis of images in the detection and diagnosis of breast cancer seminars in ultrasound. CT MRI 200; 25:411–418.
25. Erickson B.J., Bartholmai B.: Computer-aided detection and diagnosis at the start of the third millennium. J Dig Imaging 2002; 15: pp. 59-68.
26. Summers R.M.: Road maps for advancement of radiology computer-aided detection in the 21st century. Radiology 2003; 229: pp. 11-13.
27. Abe H., MacMahon H., Shiraishi J., et. al.: Computer-aided diagnosis in chest radiography. Semin Ultrasound CT MRI 2004; 25: pp. 432-437.
28. Li Q., Li F., Suzuku K., et. al.: Computer-aided diagnosis in thoracic CT. Semin Ultrasound CT MRI 2005; 26: pp. 357-363.
29. Yoshida H., Dachman A.H.: Computer-aided diagnosis for CT colonography. Semin Ultrasound CT MRI 2004; 25: pp. 404-410.
30. Doi K., Giger M.L., MacMahon H., et. al.: Computer-aided diagnosis: development of automated schemes for quantitative analysis of radiographic images. Semin Ultrasound CT MRI 1992; 13: pp. 140-152.
31. Doi K.MacMahon H.Giger M.L. et. al.Computer aided diagnosis in medical imaging.1999.ElsevierAmsterdam:
32. Doi D., MacMahon H., Katsuragawa S., et. al.: Computer-aided diagnosis in radiology: potential and pitfalls. Eur J Radiol 1999; 31: pp. 97-109.
33. Doi K.: Computer-aided diagnosis in digital chest radiography—advances in digital radiography, RSNA Categorical Course in Diagnostic Radiology Physics 2003; pp. 227-236.
34. Doi K.: Overview on research and development of computer-aided diagnosis. Semin Ultrasound CT MRI 2004; 25: pp. 404-410.
35. Doi K.: Current status and future potential of computer-aided diagnosis in medical imaging. Br J Radiol Special Issue 2005; 78: pp. S3-S19.
36. Doi K.: Diagnostic imaging over the last 50 years: research and development in medical imaging science and technology. Phys Med Biol 2006; 51: pp. R5-R27.
37. Lodwick G.S., Haun C.L., Smith W.E., et. al.: Computer diagnosis of primary bone tumor. Radiology 1963; 80: pp. 273-275.
38. Myers P.H., Nice C.M., Becker H.C., et. al.: Automated computer analysis of tracking of the vascular tree in DSA images using a double-square-box region-of-search algorithm. Proc SPIE 1986; 626: pp. 326-333.
39. Winsbarg F., Elkin M., May J., et. al.: Detection of radiographic abnormalities in mammograms by means of optical scanning and computer analysis. Radiology 1967; 89: pp. 211-215.
40. Kruger R.P., Towns J.R., Hall D.L., et. al.: Automated radiographic diagnosis via feature extraction and classification of cardiac size and shape descriptors. IEEE Trans Biomed Eng 1972; 19: pp. 174-186.
41. Kruger R.P., Thompson W.B., Turner A.F.: Computer diagnosis of pneumo-coniosis. IEEE Trans Syst Man Cybernetics 1974; 4: pp. 44-47.
42. Toriwaki J., Suenaga Y., Negoro T., et. al.: Pattern recognition of chest X-ray images. Computer Graphics Image Process 1973; 2: pp. 252-271.
43. Engle R.L.: Attempt to use computers as diagnostic aids in medical decision making: a 30-year experience. Perspect Biol Med 1992; 35: pp. 207-218.
44. Doi K.: Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Computerized Med Imaging Graphics 2007; 31: pp. 198-211.
45. Editorial: BJR review of the year – 2005. Br J Radiol 2006; 79: pp. 183-187.
46. Angelini E.D., Clatz O., Mandonnet E., et. al.: Glioma dynamics and computational models: a review of segmentation, registration, and in silico growth algorithms and their clinical applications. Curr Med Imaging Rev 2007; 3: pp. 262-276.
47. Vaidyanathan M., Clarke L.P., Velthuizen R.P., et. al.: Comparison of supervised MRI segmentation methods for tumor volume determination during therapy. Magn Reson Imaging 1995; 13: pp. 719-728.
48. Angelini E., Atif J., Delon J., et. al.: Detection of glioma evolution on longitudinal MRI studies.2007.International Symposium on Biomedical Imaging (ISBI)Arlington, VA
49. Khotanlou H., Colliot O., Atif J., et. al.: 3D brain tumor segmentation in MRI using fuzzy classification, symmetry analysis and spatially constrained deformable models. Fuzzy Sets and Systems; 2009; 160: pp. 1457-1473.
50. Dou W., Ruan S., Chen Y., et. al.: A framework of fuzzy information fusion for segmentation of brain tumor tissues on MR images. Image Vision Comp 2007; 25: pp. 164-172.
51. Duda R., Hart P.: Pattern classification and scene analysis.1973.Wiley-Interscience PublicationHolland, PA
52. Gonzalez CR, Woods ER. Digital image processing. 3rd ed; NJ: 2003
53. Staib L.H., Duncan J.S.: Parametrically deformable contour models. Comp Vision Patt Recognit Proc IEEE Comp Soc Conf 1989; pp. 98-103.
54. Gokturk S.B., Bozma H.I., Akarun L.: Automated inspection of PCBs using a novel approach. IEEE workshop on Nonlinear Signal and Image Processing 1999; pp.1865–1870
55. Bracewell R.N.: Fourier transformation and its applications.2nd ed1986.McGraw-Hill SeriesNew York
56. Garcia L. Probability and random processes for electrical engineers. 2 Sol ed. aroberts2, United States: Addison Wesley Publishing Company, 1994.
57. Mamistvalov A.G.: n-dimensional moment invariants and conceptual mathematical theory of recognition n-dimensional solids. Patt Recognit Proc 1990; 1: pp. 288-290.
58. Cambell R.J., Flynn P.J.: Eigen shapes for 3D object recognition in range data. IEEE Conference on Computer Vision and Pattern Recognition 1999;
59. Zijdenbos A.P., Dawant B.M.: Brain segmentation and white matter lesion detection in MR images. Crit Rev Biomed Eng 1994; 22: pp. 401-465.
60. Haralick R.M., Shapiro L.G.: Image segmentation techniques. Comp Vis Graph Image Proc 1985; 29: pp. 100-132.
61. Pal N.R., Pal S.K.: A review on image segmentation techniques. Pattern Recognit 1993; 26: pp. 1277-1294.
62. Bezdek J.C., Hall L.O., Clarke L.P.: Review of MR image segmentation techniques using pattern recognition. Med Phys 1993; 20: pp. 1033-1048.
63. Vaidyanathan M., Clarke L.P., Velthuizen R.P., et. al.: Comparison of supervised MRI segmentation methods for tumor volume determination during therapy. Magn Reson Imaging 1995; 13: pp. 719-728.
64. Hall L.O., Bensaid A.M., Clarke L.P., et. al.: A comparison of neural network and fuzzy clustering techniques in segmenting magnetic resonance images of the brain. IEEE Trans Neural Netw 1992; 3: pp. 672-682.
65. Sahoo P.K., Soltani S., Wong A.K.C.: A survey of thresholding techniques. Comp Vis Graph Image Proc 1988; 41: pp. 233-260.
66. Singleton H.R., Pohost G.M.: Automatic cardiac MR image segmentation using edge detection by tissue classification in pixel neighborhoods. Magn Reson Med 1997; 37: pp. 418-424.
67. Gordon L., Webber C.E., Adachi J.D., et. al.: In vivo assessment of trabecular bone structure at the distal radius from high-resolution computed tomography images. Phys Med Biol 1996; 41: pp. 495-508.
68. Polakowski W.R., Cournoyer D.A., Rogers S.K., et. al.: Computer-aided breast cancer detection and diagnosis of masses using difference of Gaussians and derivative-based feature saliency. IEEE Trans Med Imaging 1997; 16: pp. 811-819.
69. Cheng H.D., Lui Y.M., Freimanis R.I.: A novel approach to microcalcification detection using fuzzy logic technique. IEEE Trans Med Imaging 1998; 17: pp. 442-450.
70. Li D., Kallergi M., Clarke L.P., et. al.: Markov random field for tumor detection in digital mammography. IEEE Trans Med Imaging 1995; 14: pp. 565-576.
71. Lee C., Huh S., Ketter T.A., et. al.: Unsupervised connectivity-based thresholding segmentation of midsagittal brain MR images. Comp Biol Med 1998; 28: pp. 309-338.
72. Gibbs P., Buckley D.L., Blackband S.J., et. al.: Tumor volume detection from MR images by morphological segmentation. Phys Med Biol 1996; 41: pp. 2437-2446.
73. Pohlman K.A., Powell N.A., Obuchowski W.A., et. al.: Quantitative classification of breast tumors in digitized mammograms. Med Phys 1996; 23: pp. 1337-1345.
74. Manousakas N., Undrill P.E., Cameron G.G., et. al.: Split-and-merge segmentation of magnetic resonance medical images: performance evaluation and extension to three dimensions. Comp Biomed Res 1998; 31: pp. 393-412.
75. Mangin F., Frouin V., Bloch I., et. al.: From 3D magnetic resonance images to structural representations of the cortex topography using topology preserving deformations. J Math Imaging Vis 1995; 5: pp. 297-318.
76. Udupa K., Samarasekera S.: Fuzzy connectedness and object definition: theory, algorithms and applications in image segmentation. Graph Models Image Process 1996; 58: pp. 246-261.
77. Schalkoff J.: Pattern recognition: statistical, structural and neural approach.1992.Wiley & SonsNew York
78. Vannier W., Butterfield R.L., Jordan D., et. al.: Multispectral analysis of magnetic resonance images. Radiology 1985; 154: pp. 221-224.
79. Kapur T., Grimson W.E.L., Kikinis R., et. al.: Enhancedspatial priors for segmentation of magnetic resonance imagery.1998.In Proc Int Conf Med Image Comp Comp Assist IntervCambridge, MA
80. Coleman G.B., Andrews H.C.: Image segmentation by clustering. Proc IEEE 1979; 5: pp. 773-785.
81. Liang Z., MacFall J.R., Harrington D.P.: Parameter estimation and tissue segmentation from multispectral MR images. IEEE Trans Med Imaging 1994; 13: pp. 441-449.
82. Lei T., Sewchand W.: Statistical approach to X-ray CT imaging and its applications in image analysis - a new stochastic model-based image segmentation technique for X-ray CT image. IEEE Trans Med Imaging 1992; 11: pp. 62-69.
83. Jain A.K., Dubes R.C.: Algorithms for clustering data.1988.Prentice HallEnglewood Cliffs, NJ
84. Zadeh LA, Fuzzy sets. Inf Control 1965; 338–353.
85. Davenport J.W., Bezdek J.C., Hathaway R.J.: Parameter estimation for finite mixture distributions. Comp Math Appl 1988; 15: pp. 810-828.
86. Rajapakse J.C., Giedd J.N., Rapoport J.L.: Statistical approach to segmentation of single-channel cerebral MR images. IEEE Trans Med Imaging 1997; 16: pp. 176-186.
87. Pham D.L., Prince J.L.: An adaptive fuzzy c-means algorithm for image segmentation in the presence of intensity inhomogeneities. Pattern Recognit Lett 1999; 20: pp. 57-68.
88. Hebert T.J.: Fast iterative segmentation of high resolution medical images. IEEE Trans Nucl Sci 1997; 44: pp. 1363-1367.
89. Clark J.W.: Neural network modeling. Phys Med Biol 1991; 36: pp. 1259-1317.
90. Haykin S.: Neural networks: a comprehensive foundation.1994.MacmillanNew York
91. Gelenbe E., Feng Y., Krishnan K.R.R.: Neural network methods for volumetric magnetic resonance imaging of the human brain. Proc IEEE 1996; 84: pp. 1488-1496.
92. Reddick W.E., Glass J.O., Cook E.N., et. al.: Automated segmentation and classification of multispectral magnetic resonance images of brain using artificial neural networks. IEEE Trans Med Imaging 1997; 16: pp. 911-918.
93. Vilarino D.L., Brea V.M., Cabello D., et. al.: Discrete-time CNN for image segmentation by active contours. Pattern Recognit Lett 1998; 19: pp. 721-734.
94. Li S.Z.: Markov random field modeling in computer vision.1995.Springer-VerlagBerlin
95. Larie S.M., Abukmeil S.S.: Brain abnormality in schizophrenia: a systematic and quantitative review of volumetric magnetic resonance imaging studies. J Psychol 1998; 172: pp. 110-120.
96. Besag J.: On the statistical analysis of dirty pictures. CVGIP: Image Underst 1986; 57: pp. 359-372.
97. Geman S., Geman D.: Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans Pattern Anal Mach Intell 1984; 6: pp. 721-741.
98. Chen C.H., Lee G.G.: On digital mammogram segmentation and micro-calcification detection using multiresolution wavelet analysis. Graph Models Image Process 1997; 59: pp. 349-364.
99. Davatzikos C., Bryan R.N.: Using a deformable surface model to obtain a shape representation of the cortex. IEEE Trans Med Imaging 1996; 15: pp. 785-795.
100. McInerney T., Terzopoulos D.: Medical image segmentation using topologically adaptable surfaces. Lect Notes Comp Sci 1997; 1205: pp. 23-32.
101. Xu C, Pham DL, Prince JL, et al. Reconstruction of the central layer of the human cerebral cortex from MR images. In Proc Int Conf Med Image Comp Comp Assist Interv. Cambridge, MA; 1998, pp. 482–488.
102. Bardinet E., Cohen L.D., Ayache N.: A parametric deformable model to fit unstructured 3D data. Comp Vis Image Underst 1998; 71: pp. 39-54.
103. Neumann A., Lorenz C.: Statistical shape model based segmentation of medical images. Comp Med Image Graph 1998; 22: pp. 133-143.
104. Mikic I., Krucinski S., Thomas J.D.: Automatic 3-D model-based segmentation and tracking in echocardiograph sequences: active contours guided by optical low estimates. IEEE Trans Med Imaging 1995; 17: pp. 274-284.
105. Malladi R., Sethia J.A., Vemuri B.C.: Shape modeling with front propagation: a level set approach. IEEE Trans PAMI 1995; 17: pp. 158-175.
106. Cohen L.D.: On active contour models and balloons. CVGIP: Image Underst 1991; 53: pp. 211-218.
107. Caselles V., Catte F., Coll T., et. al.: A geometric model for active contours. Number Math 1993; 66: pp. 1-31.
108. Xu C., Prince J.L.: Snakes, shapes, and gradient vector flow. IEEE Trans Image Proc 1998; 7: pp. 359-369.
109. McInerney T., Terzopoulos D.: Topologically adaptable snakes.Proc Int Conf Comp Vis.1995.IEEE Comp SocCambridge, MA:pp. 840-845.
110. McInerney T., Terzopoulos D.: Deformable models in medical image analysis: a survey. Med Image Anal 1996; 1: pp. 91-108.
111. Maintz J.B.M., Viergever M.A.: A survey of medical image registration. Med Image Anal 1998; 2: pp. 1-36.
112. Talairach J., Tournoux P.: Co-planar stereotaxic atlas of the human brain. 3-dimensional proportional system: an approach to cerebral imaging.1988.ThiemeStuttgart, Germany
113. Lancaster J.L., Rainey L.H., Summerlin J.L., et. al.: Automated labeling of the human brain: a preliminary report on the development and evaluation of a forward-transform method. Hum Brain Map 1997; 5: pp. 238-242.
114. Rajarethinam N.C., Andreasen R., Cizadlo T., et. al.: Automatic atlas-based volume estimation of human brain regions from MR images. J Comp Assist Tomogr 1996; 20: pp. 98-106.
115. Collins D.L., Holmes C.J., Peters T.M.: Evans neuroanatomical segmentation. Hum Brain Mapp 1995; 3: pp. 190-208.
116. Davatzikos C.: Spatial normalization of 3D images using deformable models. J Comput Assist Tomogr 1996; 20: pp. 656-665.
117. Christensen G.E., Joshi S.C., Miller M.I.: Volumetric transformation of brain anatomy. IEEE Trans Med Imaging 1997; 16: pp. 864-877.
118. Sandor S., Leahy R.: Surface-based labeling of cortical anatomy using a de-formable atlas. IEEE Trans Med Imaging 1997; 16: pp. 41-54.
119. Aboutanos G.B., Dawant B.M.: Automatic brain segmentation and validation: image-based versus atlas-based de-formable models. SPIE Proc Med Imag 1997; 3034: pp. 299-310.
120. Thompson P., Toga A.W.: Detection, visualization and animation of abnormal anatomic structure with a probabilistic brain atlas based on random vector field transformations. Med Image Anal 1997; 1: pp. 271-294.
121. Joshi S.C., Miller M.I., Grenander U.: On the geometry and shape of brain sub-manifolds. Int J Pattern Recognit Artif Intell 1997; 11: pp. 1317-1343.
122. Pathak S.D., Grimm P.D., Chalana V., et. al.: Pubic arch detection in transrectal ultrasound guided prostate cancer therapy. IEEE Trans Med Imaging 1998; 17: pp. 762-771.
123. Bae K.T., Giger M.L., Chen C., et. al.: Automatic segmentation of liver structure in CT images. Med Phys 1993; 20: pp. 71-78.
124. Vincent L., Soille P.: Watersheds in digital spaces: an efficient algorithm based on immersion simulation. IEEE Trans Pattern Anal Mach Intell 1991; 13: pp. 583-598.
125. Sijbers J., Scheunders P., Verhoye M., et. al.: Watershed-based segmentation of 3D MR data for volume quantization. Magn Reson Imag 1997; 15: pp. 679-688.
126. Priebe C.E., Marchette D.J., Rogers G.W.: Segmentation of random fields via borrowed strength density estimation. IEEE Trans Pattern Anal Mach Intell 1997; 19: pp. 494-499.
127. Choi H.S., Hanynor D.R., Kim Y.: Partial volume tissue classification of multi-channel magnetic resonance images—a mixed model. IEEE Trans Med Imaging 1991; 10: pp. 395-407.
128. Wust P., Gellermann J., Beier J., et. al.: Evaluation of segmentation algorithms for generation of patient models in radiofrequency hyperthermia. Phys Med Biol 1998; 43: pp. 3295-3307.
129. Udupa J.K.: Fuzzy connectedness and image segmentation. Proc IEEE 2003; 91: pp. 1649-1669.
130. Hampel H., Buger K., Teipel S.J., et. al.: Core candidate neurochemical and imaging biomarkers of Alzheimer’s disease. zheimer’s Dementia 2008; 4: pp. 38-48.
131. Teipel S.J., Bokde A.L., Born C., et. al.: Morphological substrate of face matching in healthy ageing and mild cognitive impairment: a combined MRI-fMRI study. Brain 2007; 130: pp. 1745-1758.
132. Stahl R., Dietrich O., Teipel S.J., et. al.: White matter damage in Alzheimer’s disease and in mild cognitive impairment: assessment with diffusion tensor MRI using parallel imaging techniques. Radiology 2007; 243: pp. 483-492.
133. Jack C.R., Petersen R.C., Xu Y.C., et. al.: Prediction of AD with MRI-basedhippocampal volume in mild cognitive impairment. Neurology 1999; 52: pp. 1397-1403.
134. Wang P.N., Lirng J.F., Lin K.N., et. al.: Prediction of Alzheimer’s disease in mild cognitive impairment, a prospective study in Taiwan. Neurobiol Aging 2006; 27: pp. 1797-1806.
135. Ewers M., Teipel S.J., Dietrich O., et. al.: Multicenter assessment of reliability of cranial MRI. Neurobiol Aging 2006; 27: pp. 1051-1059.
136. Csernansky J.G., Wang L., Swank J., et. al.: Preclinical detection of Alzheimer’s disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage 2005; 25: pp. 783-792.
137. Ashburner J., Friston K.J.: Voxel-based morphometry: the methods. Neuroimage 2000; 11: pp. 805-821.
138. Baron J.C., Chételat G., Desgranges B., et. al.: In vivo mapping of gray matter loss with voxel-based morphometry in mild Alzheimer’s disease. Neuroimage 2001; 14: pp. 298-309.
139. Busatto G.F., Garrido G.E., Almeida O.P., et. al.: A voxel-based morphometry study of temporal lobe gray matter reductions in Alzheimer’s disease. Neurobiol Aging 2003; 24: pp. 221-223.
140. Teipel S.J., Born C., Ewers M., et. al.: Multivariate deformation-based analysis of brain atrophy to predict Alzheimer’s disease in mild cognitive impairment. Neuroimage 2007; 38: pp. 13-24.
141. Verma R., Zasharaky E.S., Ou Y., et. al.: Multiparametric tissue characterization of brain neoplasms and their recurrence using pattern classification of MR images. Acad Radiol 2008; 15: pp. 966-977.
142. Lao Z., Shen D., Liu D., et. al.: Computer-assisted segmentation of white matter lesions in 3D MR images using support vector machine. Acad Radiol 2008; 15: pp. 300-313.
143. Wang Q., Sang E., Jin R., et. al.: Segmentation of lung nodules in computed tomography images using dynamic programming and multidirection fusion techniques. Acad Radiol 2009; 16: pp. 678-6787.
144. Tokuda J., Schmitt M., Sun Y., et. al.: Lung motion and volume measurement by dynamic 3D MRI using a 128-channel receiver coil. Acad Radiol 2009; 16: pp. 22-27.
145. Zaidi H., Vees H., Wissmeye M.: Molecular PET/CT imaging–guided radiation therapy treatment planning. Acad Radiol 2009; 16: pp. 689-699.
146. Cheung M.R., Krishan K.: Interactive deformation registration of endorectal prostate MRI using ITK thin plate splines. Acad Radiol 2009; 16: pp. 351-357.
147. Fain S.B., Gonzalez-Fernandez G., Peterson E.T., et. al.: Evolution of structure function relationships in asthma using multidetector CT and hyperpolarized He-3 MRI. Acad Radiol 2008; 15: pp. 753-762.
148. Gupta V., Prakash B., Nowinski W.L.: Automatic and rapid identification of `infarct slices and hemisphere in DWI scans. Acad Radiol 2008; 15: pp. 24-39.
149. Raniga P., Bourgeat P., Fripp J., et. al.: Automated 11 C-PiB standardized uptake value ratio. Acad Radiol 2008; 15: pp. 1376-1389.
150. Bokde A.L., Lopez-Bayo P., Meindl T., et. al.: Functional connectivity of the fusiform gyrus during a face matching task in subjects with mild cognitive impairment. Brain 2006; 129: pp. 1113-1124.
151. Horwitz B., Warner B., Fitzer J., et. al.: Investigating the neural basis for functional and effective connectivity: application to fMRI. Philos Trans R Soc Lond B Biol Sci 2005; 360: pp. 1093-1108.
152. Curio G.: Linking 600-Hz spikelike EEG/MEG wavelets (sigmabursts) to cellular substrates: concepts and caveats. J Clin Neurophysiol 2000; 17: pp. 377-396.
153. Miltner W.H., Braun C., Arnold M., et. al.: Coherence of gamma-band EEG activity as a basis for associative learning. Nature 1999; 397: pp. 391-393.
154. Lantz G., Holub M., Ryding E., et. al.: Simultaneous intracranial and extracranial recording of interictal epileptiform activity in patients with therapy resistant partial epilepsy patterns of propagation and results from dipole reconstructions. Electroencephalogr Clin Neurophysiol 1996; 99: pp. 69-78.
155. Hagberg E., Bianciardi M., Maraviglia B.: Challenges for detection of neuronal currents by MRI. Magnetic Res Imaging 2006; 24: pp. 483-493.
156. Celone K.A., Calhoun V.D., Dickerson B.C., et. al.: Alterations in memory networks in mild cognitive impairment and Alzheimer’s disease: an independent component analysis. J Neurosci 2006; 26: pp. 10222-10231.
157. Greicius M.D., Srivastava G., Reiss A.L., et. al.: Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A 2004; 101: pp. 4637-4642.
158. Rombouts S.A., Barkhof F., Goekoop R., et. al.: Altered resting state networks in mild cognitive impairment and mild Alzheimer’s disease: an fMRI study. Hum Brain Mapping 2005; 26: pp. 231-239.
159. Jiang L., Song E., Xu X., et. al.: Automated detection of breast mass speculation levels and evaluation of scheme performance. Acad Radiol 2008; 15: pp. 1534-1544.
160. Sachner B., Chan H.P., Hadjiski L.M., et. al.: Multi-modality CADx: ROC study of the effect on radiologist’s accuracy in characterizing breast masses on mammograms and third ultrasound images. Acad Radiol 2009; 16: pp. 810-818.
161. Pare G., Aubry D., Lepanto L., et. al.: Evaluating PACS success: a multidimensional model. Proc IEEE Conf 2005; pp. 1-9.
162. Bick U., Lenzen H.: PACS: the silent revolution. Eur Radiol 1999; 9: pp. 1152-1160.
163. Bandon D.B., Trolliard P., Garcia A., et. al.: Building an enterprise-wide PACS for all diagnostic images. Int Congress Series 2004; 1268: pp. 279-284.
164. Ratib O., Rosset A.: Can PACS benefit from general consumer communication tools. Int Congress Series 2005; 1281: pp. 948-953.
165. Caramella D.: Is PACS research and development still necessary?. Int Congress Series 2005; 1281: pp. 11-14.