Rationale and Objectives
The Lung Image Database Consortium (LIDC) is developing a publicly available database of thoracic computed tomography (CT) scans as a medical imaging research resource to promote the development of computer-aided detection or characterization of pulmonary nodules. To obtain the best estimate of the location and spatial extent of lung nodules, expert thoracic radiologists reviewed and annotated each scan. Because a consensus panel approach was neither feasible nor desirable, a unique two-phase, multicenter data collection process was developed to allow multiple radiologists at different centers to asynchronously review and annotate each CT scan. This data collection process was also intended to capture the variability among readers.
Materials and Methods
Four radiologists reviewed each scan using the following process. In the first or “blinded” phase, each radiologist reviewed the CT scan independently. In the second or “unblinded” review phase, results from all four blinded reviews were compiled and presented to each radiologist for a second review, allowing the radiologists to review their own annotations together with the annotations of the other radiologists. The results of each radiologist’s unblinded review were compiled to form the final unblinded review. An XML-based message system was developed to communicate the results of each reading.
Results
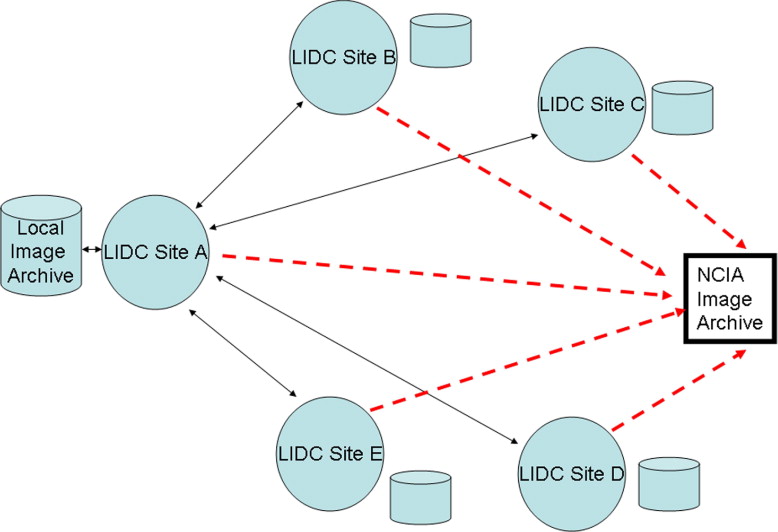
This two-phase data collection process was designed, tested, and implemented across the LIDC. More than 500 CT scans have been read and annotated using this method by four expert readers; these scans either are currently publicly available at http://ncia.nci.nih.gov or will be in the near future.
Conclusions
A unique data collection process was developed, tested, and implemented that allowed multiple readers at distributed sites to asynchronously review CT scans multiple times. This process captured the opinions of each reader regarding the location and spatial extent of lung nodules.
Computed tomography (CT) is being investigated for a variety of radiologic tasks involving lung nodules and lung malignancies. These activities include using low-dose CT as a screening tool for the early detection of lung cancer in high-risk populations ( ), evaluating the response of primary and metastatic lung lesions to various therapies ( ), and characterizing indeterminate nodules as benign or malignant ( ). Radiologists are faced with the task of both identifying and characterizing lung nodules on large, multidetector row CT scans for these applications. This has motivated interest and research into computer-aided diagnosis (CAD) methods, with several commercial systems having either already received Food and Drug Administration approval or having been submitted for approval of CAD or CAD-like systems.
To further stimulate research and development activities in this area, the National Cancer Institute (NCI) formed the Lung Image Database Consortium: the LIDC ( ). The mission of the LIDC is to 1) to develop an image database as a web-accessible international research resource for the development, training, and evaluation of CAD methods for lung cancer detection and diagnosis using CT and 2) to create this database to enable the correlation of performance of CAD methods for detection and classification of lung nodules with spatial, temporal, and pathologic ground truth. The intent of this database is to hasten advancement of lung nodule CAD research by 1) providing clinical images to investigators who might not have access to patient images and 2) creating a reference database that will support the relative comparison of different CAD systems performance, thus eliminating database composition as a source of variability in system performance ( ). This database requires the collection of an appropriate set of scans, and the creation of “truth” for each scan.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and methods
Definitions of Objects to be Marked and Annotation Requirements
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Nodules that are <3 mm but are clearly benign (ie, solidly calcified) were specifically excluded from being marked, as were nonnodules <3 mm. Each of the included categories is described below along with the annotation requirements for each. This is summarized in Table 1 .
Table 1
Summary of Categories of Objects to be Marked and the Annotation Requirements
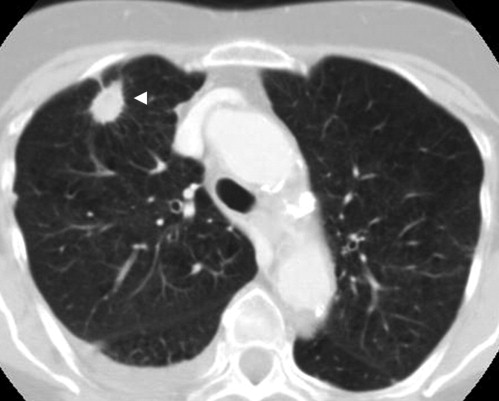
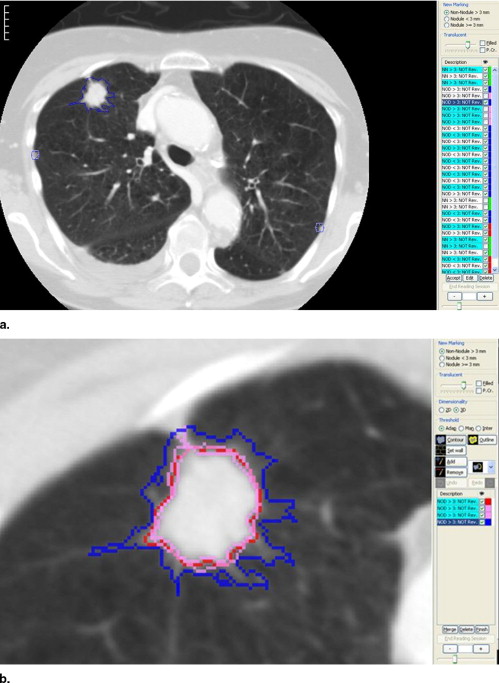
Category Annotation Subjective Assessment Nodule ≥3 mm ( Fig 1 ) Draw complete contour Yes ( Fig 7 ) Nodule <3 mm ( Fig 2 ) Mark approximate centroid None Non-nodule ≥3 mm ( Fig 3 ) Mark approximate centroid None Non-nodule <3 mm No marking None
Get Radiology Tree app to read full this article<
Nodules ≥3 mm, Regardless of Presumed Histology
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
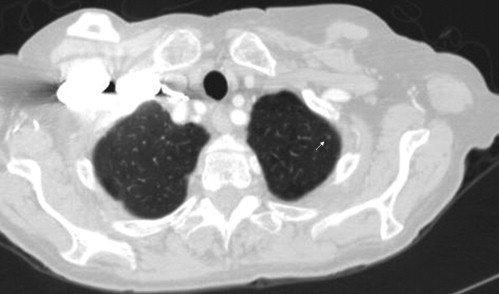
Nodules <3 mm
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
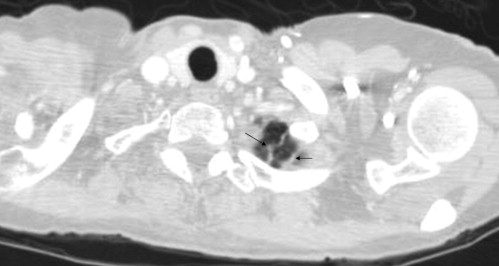
Nonnodules ≥3 mm
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Design of the Multiple Reader, Multiple Session Process
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Blinded Reading Phase
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Unblinded Reading Phase
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
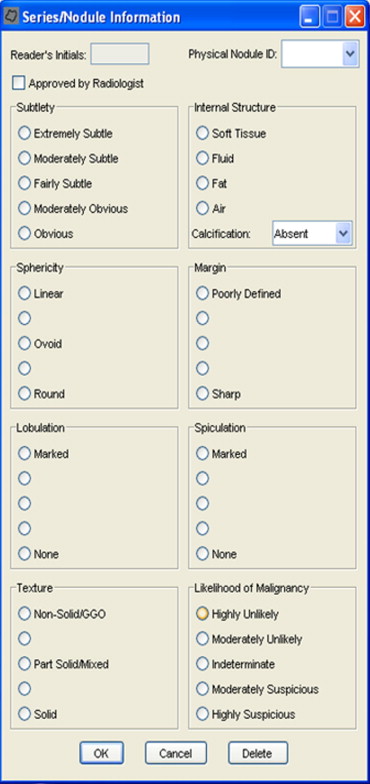
An example interface is shown in Fig 7 . This figure illustrates that each of these characteristics are rated on a five-point scale, except for internal structure and calcification. For some of the categories, descriptive terms were used for all five possible responses; for others, no descriptive terms were associated with the five possible responses. For example, a numeric value is available if the reader wants to score a lesion’s sphericity somewhere between “ovoid” and “round,” though no satisfactory descriptive term could be found.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Implementation and infrastructure
Prequisites
Get Radiology Tree app to read full this article<
Anonymization of CT Image Data
Get Radiology Tree app to read full this article<
XML-Based Specification of Annotations
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
The complete specification for the XML file is described by its schema and documentation, which can be found on the NCIA website http://ncia.nci.nih.gov/collections/ .
Get Radiology Tree app to read full this article<
Messaging System
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Software Tools for Annotation
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Results
Get Radiology Tree app to read full this article<
Conclusion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Henschke C.I., Yankelevitz D.F., Libby D.M., et. al., International Early Lung Cancer Action Program Investigators: Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med 2006; 355: pp. 1763-1771.
2. Swensen S.J., Jett J.R., Hartman T.E., et. al.: CT screening for lung cancer: five-year prospective experience. Radiology 2005; 235: pp. 259-265.
3. Zhao B., Schwartz L.H., Moskowitz C.S., et. al.: Lung cancer: computerized quantification of tumor response—initial results. Radiology 2006; 241: pp. 892-898.
4. Swensen S.J., Viggiano R.W., Midthun D.E., et. al.: Lung nodule enhancement at CT: multicenter study. Radiology 2000; 214: pp. 73-80.
5. Petkovska I., Shah S.K., McNitt-Gray M.F., et. al.: Pulmonary nodule characterization: a comparison of conventional with quantitative and visual semi-quantitative analyses using contrast enhancement maps. Eur J Radiol 2006; 59: pp. 244-252.
6. Shah S.K., McNitt-Gray M.F., Rogers S.R., et. al.: Computer aided characterization of the solitary pulmonary nodule using volumetric and contrast enhancement features. Acad Radiol 2005; 12: pp. 1310-1319.
7. Armato S.G., McLennan G., McNitt-Gray M.F., et. al., Lung Image Database Consortium Research Group: Lung image database consortium: developing a resource for the medical imaging research community. Radiology 2004; 232: pp. 739-748.
8. Clarke L.P., Croft B.Y., Staab E., et. al.: National Cancer Institute initiative: lung image database resource for imaging research. Acad Radiol 2001; 8: pp. 447-450.
9. Dodd L.E., Wagner R.F., Armato S.G., et. al., Lung Image Database Consortium Research Group: Assessment methodologies and statistical issues for computer-aided diagnosis of lung nodules in computed tomography: contemporary research topics relevant to the lung image database consortium. Acad Radiol 2004; 11: pp. 462-475.
10. Nishikawa R.M., Yarusso L.M.: Variations in measured performance of CAD schemes due to database composition and scoring protocol. Proc SPIE 1998; 3338: pp. 840-844.
11. Rogers S.R., Brown M.S., Goldin J.G., et. al.: Automated lung nodule detection in CT: nodule inclusion criteria for determining ground truth. [abstract] RSNA 2002; 225: pp. 407.
12. Wormanns D., Ludwig K., Beyer F., et. al.: Detection of pulmonary nodules at multirow-detector CT: effectiveness of double reading to improve sensitivity at standard-dose and low-dose chest CT. Eur Radiol 2005; 15: pp. 14-22.
13. Leader J.K., Warfel T.E., Fuhrman C.R., et. al.: Pulmonary nodule detection with low-dose CT of the lung: agreement among radiologists. AJR Am J Roentgenol 2005; 185: pp. 973-978.
14. Novak C.L., Qian J., Fan L., et. al.: Inter-observer variations on interpretation of multi-slice CT lung cancer screening studies, and the implications for computer-aided diagnosis. SPIE Proc 2002; 4684: pp. 68-79.
15. Meyer C.R., Johnson T.D., McLennan G., et. al.: Evaluation of lung MDCT nodule annotation across radiologists and methods. Acad Radiol 2006; 13: pp. 1254-1265.
16. Erasmus J.J., Gladish G.W., Broemeling L., et. al.: Interobserver and intraobserver variability in measurement of non-small-cell carcinoma lung lesions: implications for assessment of tumor response. J Clin Oncol 2003; 21: pp. 2574-2582.
17. Schwartz L.H., Ginsberg M.S., DeCorato D., et. al.: Evaluation of tumor measurements in oncology: use of film-based and electronic techniques. J Clin Oncol 2000; 18: pp. 2179-2184.
18. Tran L.N., Brown M.S., Goldin J.G., et. al.: Comparison of treatment response classifications between unidimensional, bidimensional, and volumetric measurements of metastatic lung lesions on chest computed tomography. Acad Radiol 2004; 11: pp. 1355-1360.
19. Judy P.F., Koelblinger C., Stuermer C., et. al.: Reliability of size measurements of patient and phantom nodules on low-dose CT, lung-cancer screening. Radiology 2002; 225: pp. 497.
20. Hopper K.D., Kasales C.J., Van Slyke M.A., et. al.: Analysis of interobserver and intraobserver variability in CT tumor measurements. AJR Am J Roentgenol 1996; 167: pp. 851-854.
21. Goodman L.R., Gulsun M., Washington L., et. al.: Inherent variability of CT lung nodule measurements in vivo using semiautomated volumetric measurements. AJR Am J Roentgenol 2006; 186: pp. 989-994.
22. Armato S.G., Starkey A., MacMahon H., et. al.: Toward a definition of lung nodule: a visual nodule library resource. [abstract] Radiology 2004; 233: pp. 708.
23. Hillman B.J., Hessel S.J., Swensson R.G., et. al.: Improving diagnostic accuracy: a comparison of interactive and Delphi consultations. Invest Radiol 1977; 12: pp. 112-115.