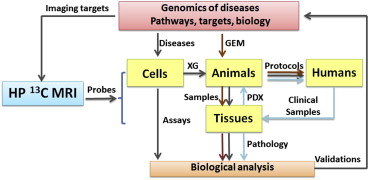
Advances in genomics are enabling integration of various -omics to reveal the complexities underneath carcinogenesis. Multivariate signaling pathways are deregulated and evolve spatially and temporally depending on the tumor microenvironment. This finding shifts the focus of cancer research from “one disease–one target and drug” to “one disease–multiple pathway targets and combinational therapy” and imposes new challenges on the imaging community in terms of imaging targets, scales and information levels. In current clinical settings, most imaging modalities assess cancer risk through alternations in anatomy, function, metabolism, cellularity, or limited molecular events. Few clinical-translatable imaging modalities are capable of detecting aberrations in signaling pathways at the level of tissue biology. An exception to this is hyperpolarized 13 C magnetic resonance spectroscopic imaging (HP 13 C MRI), which is capable of imaging the molecular signatures of special metabolic enzymes using HP 13 C-labeled substrates. HP 13 C MRI can identify multiple metabolites including intermediates and products simultaneously to allow extraction of critical parameters such as flux alterations for multiple metabolic pathways. Meanwhile, recent progress in cancer metabolism research affirms that metabolic alterations are directly controlled by signaling pathways. Thus, in vivo assessment of aberrations occurring in signaling pathways becomes feasible through HP 13 C imaging. This report briefly reviews the connections between signaling pathways and cancer metabolic phenotypes, the current status of HP 13 C MRI in assessing signal pathways, and recent advances in HP 13 C MRI techniques. Integrated with cancer genomics and animal models, HP 13 C MRI may hold high promise in exploring important issues in cancer that are linked to functionality of signaling pathways. Examples include genomic-driven therapy, intratumoral heterogeneity, and drug resistances.
Tumor growth relies on various nutrient metabolic processes such as glycolysis, lipogenesis, and glutaminolysis. Many of the important metabolites are small molecules containing carbon and nitrogen. Molecules with isotopic variants of carbon ( 13 C) and nitrogen ( 15 N) have been useful in gaining insight into the complete metabolic process by magnetic resonance spectroscopy (MRS). But 13 C MRS has rarely been used in vivo in clinical studies until recently, with the emergence of a technique called dissolution dynamic nuclear polarization. This novel hyperpolarization technique can increase 13 C signal-to-noise ratio (SNR) >10,000-fold in liquids . Such an enhancement is sufficient for collection of images with a few acquisitions or even a single scan.
Imaging modality based on this technique, the HP 13 C MRI, is expanding rapidly, primarily driven by interests of cancer metabolism. Using 13 C-labeled molecules of low molecular weight, this technique permits imaging of biochemical reactions in vivo for monitoring cancer and other diseases. Because many 13 C labeled probes have spin-lattice relaxation times (T 1 ) as short as minutes or less, an initial concern was whether this technique could have any impact on the clinical practice. To explore the clinical feasibility of HP 13 C MRI and its potential translational impact for cancer studies, the National Cancer Institute Cancer Imaging Program held a workshop at the 2010 ISMRM in Stockholm, Sweden. A consensus document (white paper) from the workshop was published in the early of 2011 . This white paper focused on the status, challenges and translational barriers to clinical applications for HP 13 C imaging and concluded that there were no significant barriers in translation to clinical utility. Recently a first-in-human study on prostate cancer was completed with HP [1- 13 C]pyruvate in 31 patients at University of California at San Francisco, demonstrating the clinical feasibility of this technology . Another clinic study with HP [1- 13 C]fumarate is under development at Cambridge University. These studies support the idea that HP 13 C MRI can become a new modality for clinical evaluation of metabolic phenotypes of cancer. Nevertheless, in the current era of cancer genomics and with the thought that all kinds of -omics are mutually attached in some as yet to be determined way, a specific question must be asked. Can HP 13 C MRI help us understand the fundamentals of cancer beyond the metabolism?
The progress in cancer genomics achieved in past decade is reshaping our strategies in evaluation and treatment of cancer. Genetic mutations, genetic instability, and microenvironment cause cancer to evolve in a complex way. Among the many suspected mutated genes found, a small group of genes have been identified that comprise a dozen essential signaling pathways to drive tumorigenesis . With these discoveries, present research paradigms are shifting from “one disease–one target and drug” to “one disease–multiple pathway targets and combinational therapies . This suggests a transition from the traditional genetic medicine, which is based on understanding the impact of single genes on disease, to a genomics-based medicine, which considers the impact of the entire genome along with environmental factors on diseases and health. This conceptual transition imposes new challenges on the imaging community where the interest is shifting to advance quantitative imaging technology to study signaling pathways in clinic setting. Recent advances in our understanding of cancer metabolism provide substantial evidence that cancer metabolism is directly linked to many signaling pathways . With these connections between cancer genomics and cancer metabolism, and with the promise and potential of HP 13 C MRI in studies of cancer metabolism, we are posed to explore the possibility of using HP 13 C MRI to assess signaling pathways in vivo, especially those critical pathways associated with tumor growth.
This report provides a brief summary of the current understanding of signaling pathways and their relationships to cancer metabolism. The unique features of HP 13 C MRI in differentiation of metabolic pathways and a summary of current applications in assessing signaling pathways and new utilities generated by the advanced imaging techniques are also presented. While HP 13 C MRI shows potential to explore various aspects of cancer metabolism, only those issues associated with manipulations and differentiations of signaling pathways will be discussed here. Because appropriate animal models offer unique systems for studying the signaling pathways, a summary of available animal models is presented. Methods of producing HP molecules with various mechanistic mechanisms are not included in this paper. Extensive reviews on this topic can be found elsewhere .
Signaling Pathways: Where are the Targets?
Comprehensive genomic sequencing efforts in the past decade have led to the completion of mutated gene maps, known as genomics landscapes, for common human cancers . In general, these landscapes contain a few high percentage mutated genes appearing as “mountains” and a much larger number of infrequently mutated genes (<5%) appearing as “hills.” About 140 of the frequently mutated genes are identified to function as “driver” of tumorigenesis, called “driver genes.” Among these driver genes, there are 64 tumor oncogenes, the genes that have high potential to cause cancer, and there are 74 tumor suppressor genes, the genes that can cause a loss of function and lead to cancer. These driver genes can be grouped into 12 signaling pathways in cell fate, cell survival, and genome maintenance, which represent three core cellular processes in tumorigenesis. Normally, oncogenes are directly targetable (i.e., it is known that 31 of the 64 oncogenes have enzymatic activities that can be targeted by drugs). Compared to the oncogenes, the tumor suppressor genes are not targetable. However, the inactivation caused by the mutation of tumor suppressor gene may activate some growth signaling at downstream that is targetable by small molecules. Therefore, multiple targeting of downstream nodes of the same signaling pathway becomes an important strategy to apply in the presence of mutated tumor suppressor genes. For typical solid tumors with 30 to 70 mutated genes, 2 to 8 of them are driver genes. Most solid tumors are dominated by the mutated tumor suppressor genes, and a few of them can contain more than one mutated oncogene. In these cases, pathway targeting or multisite simultaneous targeting become important approaches .
Metabolic Phenotypes: What are Their Linkages to Signaling Pathways?
Get Radiology Tree app to read full this article<
Metabolic Imaging: Why HP 13 C MRI?
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
HP 13 C MRI in Assessing Signaling Pathways: What Have Been Done?
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
HP 13 C MRI Techniques: What’s New?
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Assessing Signaling Pathways: What’s Next?
Potential opportunities
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Animal models
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Genomics-Driven Approach
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Conclusions
Get Radiology Tree app to read full this article<
Acknowledgment
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
References
1. Ardenkjaer-Larsen J.H., Fridlund B., Gram A., et. al.: Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci USA 2003; 100: pp. 10158-10163.
2. Kurhanewicz J., Vigneron D.B., Brindle K., et. al.: Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia 2011; 13: pp. 81-97.
3. Nelson S.J., Kurhanewicz J., Vigneron D.B., et. al.: Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(13)C]pyruvate. Sci Transl Med 2013; 5: pp. 198-208.
4. Vogelstein B., Papadopoulos N., Velculescu V.E., et. al.: Cancer genome landscapes. Science 2013; 339: pp. 1546-1558.
5. Yap T.A., Omlin A., de Bono J.S.: Development of therapeutic combinations targeting major cancer signaling pathways. J Clin Oncol 2013; 31: pp. 1592-1605.
6. Garraway L.A.: Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol 2013; 31: pp. 1806-1814.
7. Garay J.P., Gray J.W.: Omics and therapy: a basis for precision medicine. Mol Oncol 2012; 6: pp. 128-139.
8. Jones R.G., Thompson C.B.: Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev 2009; 23: pp. 537-548.
9. Ward P.S., Thompson C.B.: Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 2012; 21: pp. 297-308.
10. DeBerardinis R.J., Thompson C.B.: Cellular metabolism and disease: what do metabolic outliers teach us?. Cell 2012; 148: pp. 1132-1144.
11. Kroemer G., Pouyssegur J.: Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 2008; 13: pp. 472-482.
12. Cairns R.A., Harris I.S., Mak T.W.: Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11: pp. 85-95.
13. Brindle K.: Watching tumours gasp and die with MRI: the promise of hyperpolarised 13C MR spectroscopic imaging. Br J Radiol 2012; 85: pp. 697-708.
14. Nelson S.J., Ozhinsky E., Li Y., et. al.: Strategies for rapid in vivo 1H and hyperpolarized 13C MR spectroscopic imaging. J Magn Reson 2013; 229: pp. 187-197.
15. Dang C.V.: MYC on the path to cancer. Cell 2012; 149: pp. 22-35.
16. Linehan W.M., Srinivasan R., Schmidt L.S.: The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol 2010; 7: pp. 277-285.
17. Condeelis J., Weissleder R.: In vivo imaging in cancer. Cold Spring Harb Perspect Biol 2010; 2: pp. a003848.
18. James M.L., Gambhir S.S.: A molecular imaging primer: modalities, imaging agents, and applications. Physiol Rev 2012; 92: pp. 897-965.
19. Wistuba I.I., Gelovani J.G., Jacoby J.J., et. al.: Methodological and practical challenges for personalized cancer therapies. Nat Rev Clin Oncol 2011; 8: pp. 135-141.
20. Dafni H., Larson P.E., Hu S., et. al.: Hyperpolarized 13C spectroscopic imaging informs on hypoxia-inducible factor-1 and myc activity downstream of platelet-derived growth factor receptor. Cancer Res 2010; 70: pp. 7400-7410.
21. Chaumeil M.M., Ozawa T., Park I., et. al.: Hyperpolarized 13C MR spectroscopic imaging can be used to monitor everolimus treatment in vivo in an orthotopic rodent model of glioblastoma. NeuroImage 2012; 59: pp. 193-201.
22. Lodi A., Woods S.M., Ronen S.M.: Treatment with the MEK inhibitor U0126 induces decreased hyperpolarized pyruvate to lactate conversion in breast, but not prostate, cancer cells. NMR Biomed 2013; 26: pp. 299-306.
23. Hu S., Balakrishnan A., Bok R.A., et. al.: 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab 2011; 14: pp. 131-142.
24. Dutta P., Le A., Vander Jagt D.L., et. al.: Evaluation of LDH-A and glutaminase inhibition in vivo by hyperpolarized 13C-pyruvate magnetic resonance spectroscopy of tumors. Cancer Res 2013; 73: pp. 4190-4195.
25. Bohndiek S.E., Kettunen M.I., Hu D.E., et. al.: Hyperpolarized (13)C spectroscopy detects early changes in tumor vasculature and metabolism after VEGF neutralization. Cancer Res 2012; 72: pp. 854-864.
26. Ardenkjaer-Larsen J.H., Leach A.M., Clarke N., et. al.: Dynamic nuclear polarization polarizer for sterile use intent. NMR Biomed 2011; 24: pp. 927-932.
27. Hu S., Larson P.E., Vancriekinge M., et. al.: Rapid sequential injections of hyperpolarized [1-(1)(3)C]pyruvate in vivo using a sub-kelvin, multi-sample DNP polarizer. Magn Reson Imaging 2013; 31: pp. 490-496.
28. Wilson D.M., Keshari K.R., Larson P.E., et. al.: Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo. J Magn Reson 2010; 205: pp. 141-147.
29. Meier S., Karlsson M., Jensen P.R., et. al.: Metabolic pathway visualization in living yeast by DNP-NMR. Mol Biosyst 2011; 7: pp. 2834-2836.
30. Ohliger M.A., Larson P.E., Bok R.A., et. al.: Combined parallel and partial fourier MR reconstruction for accelerated 8-channel hyperpolarized carbon-13 in vivo magnetic resonance spectroscopic imaging (MRSI). J Magn Reson Imaging 2013; 38: pp. 701-713.
31. Keshari K.R., Sriram R., Van Criekinge M., et. al.: Metabolic reprogramming and validation of hyperpolarized (13) C lactate as a prostate cancer biomarker using a human prostate tissue slice culture bioreactor. Prostate 2013; 73: pp. 1171-1181.
32. Li L.Z., Kadlececk S., Xu H.N., et. al.: Ratiometric analysis in hyperpolarized NMR (I): test of the two-site exchange model and the quantification of reaction rate constants. NMR Biomed 2013; 26: pp. 1308-1320.
33. Swisher C.L., Larson P.E., Kruttwig K., et. al.: Quantitative measurement of cancer metabolism using stimulated echo hyperpolarized carbon-13 MRS. Magn Reson Med 2013, Feb 14; [Epub ahead of print]
34. Heiser L.M., Sadanandam A., Kuo W.L., et. al.: Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A 2012; 109: pp. 2724-2729.
35. Woodcock J., Griffin J.P., Behrman R.E.: Development of novel combination therapies. N Engl J Med 2011; 364: pp. 985-987.
36. Ding L., Ley T.J., Larson D.E., et. al.: Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012; 481: pp. 506-510.
37. Gerlinger M., Rowan A.J., Horswell S., et. al.: Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: pp. 883-892.
38. Marusyk A., Polyak K.: Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 2010; 1805: pp. 105-117.
39. Engelman J.A.: Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 2009; 9: pp. 550-562.
40. Grinde M.T., Moestue S.A., Borgan E., et. al.: 13C high-resolution-magic angle spinning MRS reveals differences in glucose metabolism between two breast cancer xenograft models with different gene expression patterns. NMR Biomed 2011; 24: pp. 1243-1252.
41. Perou C.M., Borresen-Dale A.L.: Systems biology and genomics of breast cancer. Cold Spring Harb Perspect Biol 2011; 3:
42. Van Dyke T., Jacks T.: Cancer modeling in the modern era: progress and challenges. Cell 2002; 108: pp. 135-144.
43. Cheon D.J., Orsulic S.: Mouse models of cancer. Annu Rev Pathol 2011; 6: pp. 95-119.
44. Tentler J.J., Tan A.C., Weekes C.D., et. al.: Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 2012; 9: pp. 338-350.
45. Sharpless N.E., Depinho R.A.: The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov 2006; 5: pp. 741-754.
46. Singh M., Murriel C.L., Johnson L.: Genetically engineered mouse models: closing the gap between preclinical data and trial outcomes. Cancer Res 2012; 72: pp. 2695-2700.
47. Nardella C., Lunardi A., Patnaik A., et. al.: The APL paradigm and the “co-clinical trial” project. Cancer Discov 2011; 1: pp. 108-116.
48. Chen Z., Cheng K., Walton Z., et. al.: A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012; 483: pp. 613-617.