Rationale and Objectives
To point out the problems with Cohen kappa statistic and to explore alternative metrics to determine interobserver agreement on lesion detection when locations are not prespecified.
Materials and Methods
Use of kappa and two alternative methods, namely index of specific agreement (ISA) and modified kappa, for measuring interobserver agreement on the location of detected lesions are presented. These indices of agreement are illustrated by application to a retrospective multireader study in which nine readers detected and scored prostate cancer lesions in 163 consecutive patients ( n = 110 cases, n = 53 controls) using the guideline of Prostate Imaging Reporting and Data System version 2 on multiparametric magnetic resonance imaging.
Results
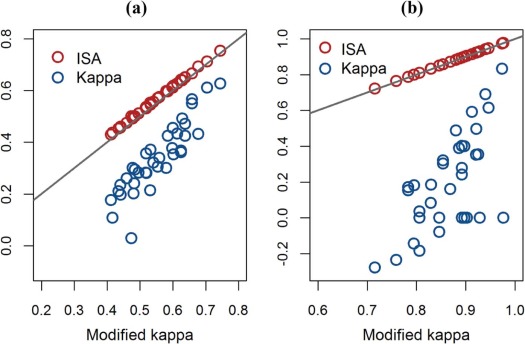
The proposed modified kappa, which properly corrects for the amount of agreement by chance, is shown to be approximately equivalent to the ISA. In the prostate cancer data, average kappa, modified kappa, and ISA equaled 30%, 55%, and 57%, respectively, for all lesions and 20%, 87%, and 87%, respectively, for index lesions.
Conclusions
The application of kappa could result in a substantial downward bias in reader agreement on lesion detection when locations are not prespecified. ISA is recommended for assessment of reader agreement on lesion detection.
Introduction
Imaging plays an integral role in cancer lesion detection and characterization in oncology practice. As subjective imaging features often require a high level of expertise, imaging modalities must be shown to have acceptable reproducibility before they can be widely used as diagnostic tools and assist in treatment decisions. For this purpose, imaging techniques have been evaluated for interobserver agreement in multireader studies. A common design used in these studies is to have multiple readers normally blinded to clinical and pathologic outcomes score each object on an image independently, using a dichotomized classification or ordinal scale to characterize the likelihood of clinical significance . The imaging objects might be lesions previously identified, sectors of an organ, or multiple anatomical districts .
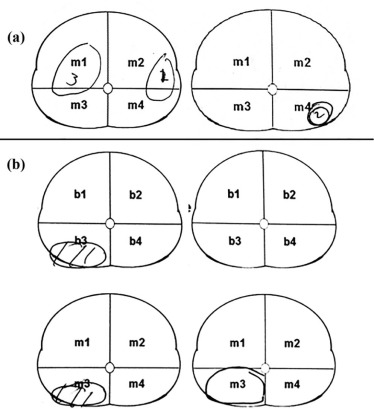
Recently, an alternative design has been used in multireader studies in which readers are asked to identify and score lesions on images . Two types of interobserver agreement can be simultaneously assessed in studies implemented under this design: (1) lesion detection and (2) scoring of identified lesions. Agreement on scoring of identified lesions can be calculated by kappa. Determination of reader agreement on lesion detection, however, is challenging because lesions can appear anywhere on an image and readers vary in interpretation of lesion location. For example, two readers might map distinct lesions to the same sector or map the same lesion to different sectors as demonstrated in Figure 1 for the sector map of prostate, resulting in false-positive (FP) and false-negative agreement, respectively.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Materials and Methods
Prostate Cancer Multiparametric MRI Multireader Study
Get Radiology Tree app to read full this article<
Kappa Statistic
Get Radiology Tree app to read full this article<
TABLE 1
Lesion Detection Tabulation of Two Readers
Reader 2 Undetected Total Detected Reader 1 Detected_a__b__a_ + b Undetected_c__d__c_ + d Total_a_ + c__b + d__a + b + c + d
Get Radiology Tree app to read full this article<
Index of Specific Agreement (ISA)
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Reader Agreement on Absence of Lesions
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
TABLE 2
Tabulation of Number of Regions of Two Readers with Respect to Lesion Presence, Where K denotes the Number of Mutually Exclusive Regions in Each Image and n Denotes the Total Number of Images Read by Both Readers
Reader 2 Absence Total Presence Reader 1 Presence_a__b__a_ + b Absence_c__nK-(a_ + b + c)__nK-(a + b ) Total_a_ + c__nK-(a + c)__nK
Get Radiology Tree app to read full this article<
Relationship between ISA and Modified Kappa
Get Radiology Tree app to read full this article<
Complementary Measures of Agreement
Get Radiology Tree app to read full this article<
Simulation Study
Get Radiology Tree app to read full this article<
Analysis
Get Radiology Tree app to read full this article<
Results
Prostate Cancer mpMRI Imaging Multireader Study
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
TABLE 3
Index of Specific Agreement and 95% Bootstrap Confidence Interval on the Location of Detected Lesions for Nine Readers
Overall H-H H-ML ML-ML All lesions 0.57 (0.50, 0.63) 0.66 (0.56, 0.75) 0.59 (0.51, 0.65) 0.53 (0.45, 0.60) Index lesions 0.87 (0.80, 0.92) 0.90 (0.83, 0.97) 0.88 (0.82, 0.93) 0.84 (0.75, 0.91)
H-H: Agreement between highly experienced readers.
H-ML: Agreement between highly and moderately + low experienced readers.
ML-ML: Agreement among moderately and low experienced readers.
TABLE 4
Proportions of Agreement in True Positive (TP) and False Positive (FP) on Lesion Detection and 95% Bootstrap Confidence Intervals
Overall H-H H-ML ML-ML All lesions TP 0.59 (0.51–0.69) 0.64 (0.53,0.77) 0.60 (0.51–0.70) 0.58 (0.49,0.68) FP 0.11 (0.06–0.15) 0.06 (0,0.22) 0.10 (0.05–0.16) 0.12 (0.08,0.16) Index lesions TP 0.78 (0.67–0.86) 0.83 (0.70–0.94) 0.80 (0.71–0.88) 0.73 (0.62–0.83)
Get Radiology Tree app to read full this article<
Simulation Study
Get Radiology Tree app to read full this article<
TABLE 5
Results of the Two-reader Simulation Study
Number of Images True ISA Monte Carlo Mean of Kappa Monte Carlo Mean of ISA Monte Carlo Mean of Modified ISA K = 20 K = 40 K = 60 50 0.67 0.40 0.67 0.64 0.65 0.66 100 0.67 0.40 0.67 0.64 0.65 0.66
ISA, index of specific agreement.
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Acknowledgments
Get Radiology Tree app to read full this article<
Appendix
Formula for Modified Kappa
Get Radiology Tree app to read full this article<
Modifiedkappa=nK−b−cnK−(a+b)(a+c)n2K2−[1−a+bnK][1−a+cnK]1−(a+b)(a+c)n2K2−[1−a+bnK][1−a+cnK]=2anK−2(a+b)(a+c)n2K22a+b+cnK−2(a+b)(a+c)n2K2. Modified
kappa
=
n
K
−
b
−
c
n
K
−
(
a
+
b
)
(
a
+
c
)
n
2
K
2
−
[
1
−
a
+
b
n
K
]
[
1
−
a
+
c
n
K
]
1
−
(
a
+
b
)
(
a
+
c
)
n
2
K
2
−
[
1
−
a
+
b
n
K
]
[
1
−
a
+
c
n
K
]
=
2
a
n
K
−
2
(
a
+
b
)
(
a
+
c
)
n
2
K
2
2
a
+
b
+
c
n
K
−
2
(
a
+
b
)
(
a
+
c
)
n
2
K
2
.
Get Radiology Tree app to read full this article<
Relationship between modified kappa and ISA
Get Radiology Tree app to read full this article<
Modifiedkappa=2p11K−2p1+p+1K2p1++p+1K−2p1+p+1K2=2p11−2p1+p+1Kp1++p+1−2p1+p+1K. Modified
kappa
=
2
p
11
K
−
2
p
1
+
p
+
1
K
2
p
1
+
+
p
+
1
K
−
2
p
1
+
p
+
1
K
2
=
2
p
11
−
2
p
1
+
p
+
1
K
p
1
+
+
p
+
1
−
2
p
1
+
p
+
1
K
.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Relationship between kappa and ISA
Get Radiology Tree app to read full this article<
kappa=a+d−(a+b)(a+c)−(b+d)(c+d)a+b+c+d kappa
=
a
+
d
−
(
a
+
b
)
(
a
+
c
)
−
(
b
+
d
)
(
c
+
d
)
a
+
b
+
c
+
d
≤2a+d−(a+b)(a+c)−(b+d)(c+d)2a+b+c+d ≤
2
a
+
d
−
(
a
+
b
)
(
a
+
c
)
−
(
b
+
d
)
(
c
+
d
)
2
a
+
b
+
c
+
d
≤2a2a+b+c+d ≤
2
a
2
a
+
b
+
c
+
d
2a2a+b+c=ISA. 2
a
2
a
+
b
+
c
=
I
S
A
.
Get Radiology Tree app to read full this article<
Supplementary Data
Get Radiology Tree app to read full this article<
Video 1
Get Radiology Tree app to read full this article<
References
1. Muller B.G., Shih J.H., Sankineni S., et. al.: Prostate cancer: interobserver agreement and accuracy with the revised prostate imaging reporting and data system at multiparametric MR imaging. Radiology 2015; 277: pp. 741-750.
2. Kasel-Seibert M., Lehmann T., Aschenbach R., et. al.: Assessment of PI-RADS v2 for the detection of prostate cancer. Eur J Radiol 2016; 85: pp. 726-731.
3. Zacchino M., Bonaffini P.A., Corso A., et. al.: Inter-observer agreement for the evaluation of bone involmement on while body low dose computed tomograph (WBLDCT) in multiple myeloma (MM). Eur Radiol 2015; 25: pp. 3382-3389.
4. Gollub M.J., Lakhman Y., McGinty K., et. al.: Does gadolinium-based contrast material improve diagnostic accuracy of local invasion in rectal cancer MRI? A multireader study. AJR Am J Roentgenol 2015; 204: pp. W160-W167.
5. Mariscotti G., Durando M., Houssami N., et. al.: Digital breast tomosynthesis as an adjunct to digital mammography for detecting and characterizing invasive lobular cancers: a multi-reader study. Clin Radiol 2016; 71: pp. 889-895.
6. Greer M.D., Brown A.M., Shih J.H., et. al.: Accuracy and agreement of PIRADSv2 for prostate cancer mpMRI: a multi-reader study. J Magn Reson Imaging 2017; 45: pp. 579-585.
7. Greer M.D., Shih J.H., Lay N., et. al.: Validation of the dominant sequence paradigm and role of DCE in PIRADSv2. Radiology 2017; 285: pp. 859-869.
8. Chen F., Cen S., Palmer S.: Application of prostate imaging reporting and data system version 2 (PI-RADS v2): interobserver agreement and positive predictive value for localization of intermediate- and high-grade prostate cancers on multiparametric magnetic resonance imaging. Acad Radiol 2017; 24: pp. 1101-1106.
9. Greer M.D., Choyke P.L., Turkbey B.: PI-RADSv2: How we do it. J Magn Reson Imaging 2017; 46: pp. 11-23.
10. Radiology ACo : Prostate imaging reporting and data system version 2.0.2015.
11. Viera A.J., Garrett J.M.: Understanding interobserver agreement: the kappa statistic. Fam Med 2005; 37: pp. 360-363.
12. Monseruda R.A., Leemans R.: Comparing global vegetation maps with the Kappa statistic. Ecol Modell 1992; 62: pp. 275-293.
13. Rasheed K., Rabinowitz Y.S., Remba D., et. al.: Interobserver and intraobserver reliability of a classification scheme for corneal topographic patterns. Br J Ophthalmol 1998; 82: pp. 1401-1406.
14. Rasheed K., Rabinowitz Y.S., Remba D., et. al.: Interobserver and intraobserver reliability of a classification scheme for corneal topographic patterns. J Opthalmology 1998; 82: pp. 1401-1406.
15. Visser H., de Nijs T.: The map comparison kit. Environ Model Softw 2006; 21: pp. 346-358.
16. Fleiss J.L.: Statistical methods for rates and proportions.2nd ed.1981.John WileyNew York
17. Cicchetti D.V., Feinstein A.R.: High agreement but low kappa: II. Resolving the paradoxes. J Clin Epidemiol 1990; 43: pp. 551-558.
18. Shoukri M.M.: Measures of interobserver agreement and reliability.2nd ed.2011.CRC Press, Taylor & Francis GroupNew York, NY
19. Pontius R.G., Millones M.: Death to kappa: birth of quantity disagreement and allocation disagreement for accuracy assessment. Int J Remote Sens 2011; 32: pp. 4407-4429.
20. Carpentier M., Combescure C., Merlini L., et. al.: Kappa statistic to measure agreement beyond chance in free-response assessments. BMC Med Res Methodol 2017; 17: pp. 62.
21. Steenbergen P., Haustermans K., Lerut E., et. al.: Prostate tumor delineation using multiparametric magnetic resonance imaging: Inter-observer variability and pathology validation. Radiat Oncol 2015; 115: pp. 186-190.
22. Zou K.H., Warfield S.K., Bharatha A., et. al.: Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 2004; 11: pp. 178-189.
23. He X., Frey E.: ROC, LROC, FROC, AFROC: an alphabet soup. Am Coll Radiol 2009; 6: pp. 652-655.