Rationale and Objectives
The goal of many multiple-observer computer-aided detection (CADe) studies is to estimate the change in observers’ diagnostic performance with CADe from their unaided performance. A key issue in these studies is the method for estimating the observers’ unaided performance. The crossover design is considered the most valid. The sequential design takes less time and is less expensive but may be biased. We conducted a study to investigate the differences between these two designs.
Materials and Methods
Data from two large CADe studies using both types of unaided reads were analyzed. The first study involved three radiologists examining the chest x-rays of 200 patients for lung nodules. The second study involved 19 observers interpreting the computed tomography colonography images of 100 patients for polyps. Observers’ sensitivity, specificity, and receiver operating characteristic areas were estimated while unaided in both designs and compared to their accuracy with CADe. Bias, inter-observer variability, and correlations between unaided and aided results were assessed.
Results
Observers tend to perform better while unaided in the sequential design than while unaided in the crossover design, but the differences are small. The inter-observer variability is larger in the sequential design. The correlations between unaided and aided results are larger in the sequential design. 95% CIs for the change with CADe are narrower with the sequential design.
Conclusion
The estimated effect of CADe on observer performance is similar regardless of the study design. Use of the sequential design may save investigators time and resources.
Observer studies comparing the diagnostic performance without and with the use of computer-aided detection (CADe) are critical in verifying the clinical utility of CADe even if excellent stand-alone performance is proven . Numerous observer studies have been published in which CADe is used to help detect lesions in various clinical settings: breast lesions on mammography, lung nodules on plain films, pulmonary emboli on computed tomography (CT), and colonic polyps on CT colonography .
A key issue in the design of these CADe studies is the method for estimating the observers’ performance without CADe (ie, unaided performance). There are three study design methods that have been used in prior studies: historical control, crossover control, and sequential control ( Fig 1 ).
Figure 1
Comparison of the three common CADe study designs.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Methods
Clinical Studies
Get Radiology Tree app to read full this article<
Table 1
Inclusion Criteria for Two Studies
Study #1: CADe for Chest X-ray Study #2: CADe for CTC
Reference standard An expert observer reviewed the CT and/or PET images, and pathology results if available. Based on this review, the expert established ground truth about the presence or absence of actionable nodules, and determined the size, shape, and location (specified as one of 10 predefined lung regions) of all actionable nodules. The expert was not blinded to the original CT or PET interpretations but was blinded to the CADe marks. Two expert observers independently reviewed the optical colonoscopy and CTC images. When the experts did not agree, a third expert reviewed the images. This process was used to establish ground truth about the presence or absence of actionable polyps and the size, morphology, and spatial coordinates of all actionable polyps. The experts were not blinded to the original CTC or optical colonoscopy interpretations, but were blinded to the CADe marks.
CT, computed tomography; CADe, computer-aided detection; CTC, CT colonography; PET, positron emission tomography.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Statistical Methods
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Results
Assessment of Bias
Get Radiology Tree app to read full this article<
Table 2
Average of Observers’ Diagnostic Accuracy (Standard Error)
Unaided Accuracy (SE of difference)
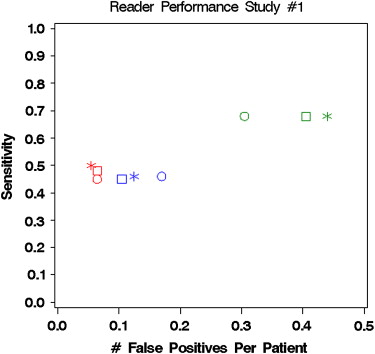
95% CIs for Difference Crossover Sequential CAD CADe - Crossover CADe – Sequential Study 1 Patient ROC area 0.727 (0.022) 0.737 (0.026) 0.740 (0.033) (0.02404)
−0.091, 0.116 (.00867)
−0.035, 0.040 Patient sensitivity 0.530 (0.082) 0.537 (0.079) 0.547 (0.075) (0.01653)
−0.055, 0.088 (0.00574)
−0.015, 0.035 Patient specificity 0.893 (0.056) 0.900 (0.076) 0.893 (0.083) (0.03033)
−0.131, 0.131 (0.01049)
−0.039, 0.052 Study 2 Segment ROC area 0.737 (0.029) ∗ 0.743 (0.029) ∗ 0.758 (0.029) (0.008187)
0.004, 0.039 (0.00400)
0.007, 0.024 Patient ROC area 0.711 (0.030) 0.716 (0.030) ∗ 0.727 (0.030) (0.00854)
−0.002, 0.034 (0.00516)
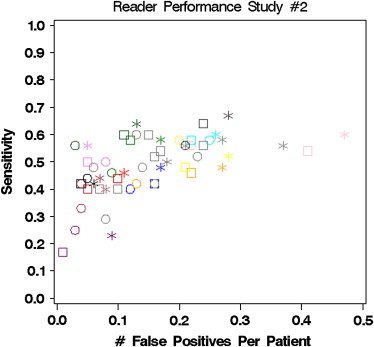
0.0003, 0.022 Segment sensitivity 0.465 (0.057) ∗ 0.487 (0.057) ∗ 0.517 (0.057) (0.019324)
0.012, 0.093 (0.007401)
0.015, 0.046 Patient sensitivity 0.466 (0.055) ∗ 0.487 (0.055) ∗ 0.521 (0.055) (0.019443)
0.015, 0.097 (0.009281)
0.015, 0.054 Segment specificity 0.984 (0.004) ∗ 0.981 (0.004) ∗ 0.975 (0.004) (0.002499)
−0.014, −0.004 (0.001523)
−0.009, −0.003 Patient specificity 0.929 (0.020) 0.917
(0.023) ∗ 0.904
(0.022) (0.012089)
−0.051, 0.0002 (0.005545)
−0.025, −0.002
CADe, computer-aided detection; ROC, receiver operating characteristic; crossover, crossover study design.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Assessment of Inter-Observer Variability
Get Radiology Tree app to read full this article<
Table 3
Inter-Observer Variances ∗
Crossover Sequential Study #1 Patient - ROC area 0.0004 (NA) 0.0010 (0.0005) Patient - sensitivity 0.0169 (0.0156) 0.0156 (0.0142) Patient - specificity 0.0092 (0.0077) 0.0171 (0.0161) Study #2 Segment - ROC area 0.0028 (0.0026) 0.0032 (0.0027) Patient - ROC area 0.0023 (0.0015) 0.0027 (0.0022) Segment - sensitivity 0.0107 (0.0075) 0.0123 (0.0097) Patient - sensitivity 0.0099 (0.0071) 0.0113 (0.0084) Segment - specificity 0.0001 (0.0001) 0.0002 (0.0002) Patient - specificity 0.0028 (0.0013) 0.0039 (0.0032)
ROC, receiver operating characteristic; NA, the model-based estimate was negative.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Assessment of Correlation between Unaided and Aided Results
Get Radiology Tree app to read full this article<
Table 4
Correlations Between Results With vs. Without CADe
Crossover vs. CADe Sequential vs. CADe Study #1 Correlation between scores ∗ 0.64 0.89 Correlation between ROC areas † 0.50 1.0 Study #2 Correlation between scores ∗ 0.71 0.94 Correlation between ROC areas ∗ 0.83 0.95
ROC, receiver operating characteristic; CADe, computer-aided detection.
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Discussion
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Get Radiology Tree app to read full this article<
Acknowledgment
Get Radiology Tree app to read full this article<
References
1. Dodd L.E., Wagner R.F., Armato S.G., et. al.: Lung Image Database Consortium Research Group. Assessment methodologies and statistical issues for computer-aided diagnosis of lung nodules in computed tomography: contemporary research topics relevant to the lung image database consortium. Acad Radiol 2004; 11: pp. 462-475.
2. Wagner R.F., Metz C.E., Campbell G.: Assessment of medical imaging systems and computer aids: a tutorial review. Acad Radiol 2007; 14: pp. 723-748.
3. Das M., Muhlenbruch G., Mahnken A.H., et. al.: Small pulmonary nodules: effect of two computer-aided detection systems on radiologists performance. Radiology 2006; 241: pp. 564-571.
4. Halligan S., Altman D.G., Mallett S., et. al.: Computed tomographic colonography: assessment of radiologists performance with and without computer-aided detection. Gastroenterology 2006; 131: pp. 1690-1699.
5. Fenton J.J., Taplin S.H., Carney P.A., et. al.: Influence of computer-aided detection on performance of screening mammography. N Engl J Med 2007; 356: pp. 1399-1409.
6. Birdwell R.L., Bandodkar P., Ikeda D.M.: Computer-aided detection with screening mammography in a university hospital setting. Radiology 2005; 236: pp. 451-457.
7. Gur D., Sumkin J.H., Rockette H.E., et. al.: Changes in breast cancer detection and mammography recall rates after the introduction of a computer-aided detection system. J Natl Cancer Inst 2004; 96: pp. 185-190.
8. Cupples T.E., Cunningham J.E., Reynolds J.C.: Impact of computer-aided detection in a regional screening mammography program. AJR Am J Radiol 2005; 185: pp. 944-950.
9. Rutter C.M., Taplin S.: Assessing mammographers’ accuracy: a comparison of clinical and test performance. J Clin Epidemiol 2000; 53: pp. 443-450.
10. Gur D.: ROC-type assessment of medical imaging and CAD technologies. Acad Radiol 2003; 10: pp. 402-403.
11. Gur D.: Objectively measuring and comparing performance levels of diagnostic imaging systems and practices. Acad Radiol 2007; 14: pp. 641-642.
12. Draft Guidance for Industry and FDA Staff, page 12. Available at www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm187277.htm . Accessed February 16, 2010.
13. Zhou X.H., Obuchowski N.A., McClish D.L.: Statistical methods in diagnostic medicine.2002.Wiley and Sons, IncNew York
14. Zalis M.E., Barish M.A., Choi J.R., et. al.: CT colonography reporting and data system: a consensus proposal. Radiology 2005; 236: pp. 3-9.
15. Obuchowski N.A., Mazzone P.J., Dachman A.H.: Bias, underestimation of risk and loss of statistical power in patient-level analyses of lesion detection. Eur Radiol 2009; 20: pp. 584-594.
16. DeLong E., DeLong D., Clarke-Pearson D.: Comparing areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: pp. 837-845.
17. Obuchowski N.A.: Nonparametric analysis of clustered ROC curve data. Biometrics 1997; 53: pp. 170-180.
18. Obuchowski N.A., Rockette H.E.: Hypothesis testing of the diagnostic accuracy for multiple diagnostic tests: an ANOVA approach with dependent observations. Commun Stat Simulation Comput 1995; 24: pp. 285-308.